
Saccharomyces 20S RNA narnavirus (ScNV-20S)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Amabiliviricetes; Wolframvirales; Narnaviridae; Narnavirus
Average proteome isoelectric point is 9.25
Get precalculated fractions of proteins
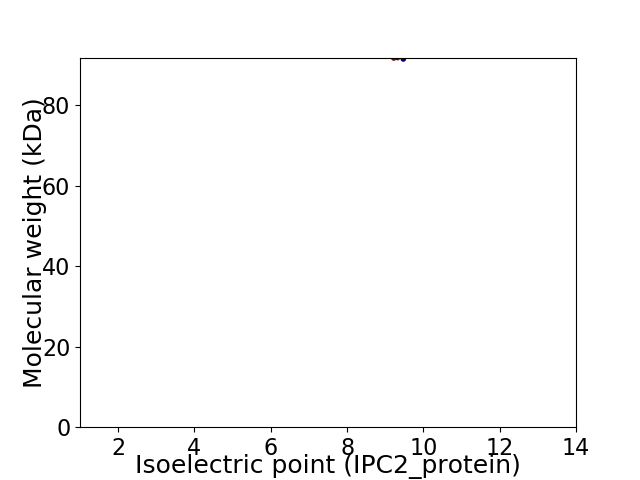
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P25328|RDRP_SCV20 RNA-directed RNA polymerase OS=Saccharomyces 20S RNA narnavirus OX=186772 PE=1 SV=1
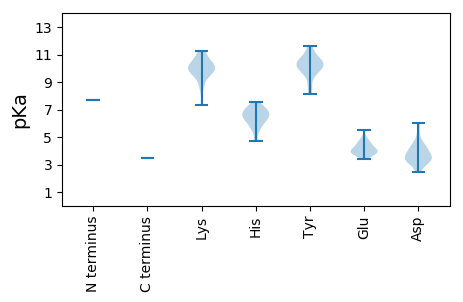
MM1 pKa = 7.68KK2 pKa = 10.46EE3 pKa = 3.87PVDD6 pKa = 4.16CRR8 pKa = 11.84LSTPAGFSGTVPPPGRR24 pKa = 11.84TKK26 pKa = 10.44AARR29 pKa = 11.84PGTIPVRR36 pKa = 11.84RR37 pKa = 11.84SRR39 pKa = 11.84GSASALPGKK48 pKa = 10.24IYY50 pKa = 10.22GWSRR54 pKa = 11.84RR55 pKa = 11.84QRR57 pKa = 11.84DD58 pKa = 3.69RR59 pKa = 11.84FAMLLSSFDD68 pKa = 3.67AALAAYY74 pKa = 9.68SGVVVSRR81 pKa = 11.84GTRR84 pKa = 11.84SLPPSLRR91 pKa = 11.84LFRR94 pKa = 11.84AMTRR98 pKa = 11.84KK99 pKa = 8.74WLSVTARR106 pKa = 11.84GNGVEE111 pKa = 4.18FAIASAKK118 pKa = 9.88EE119 pKa = 3.6FSAACRR125 pKa = 11.84AGWISGTVPDD135 pKa = 4.5HH136 pKa = 7.26FFMKK140 pKa = 10.09WLPEE144 pKa = 3.67PVRR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 10.1SGLWAQLSFIGRR161 pKa = 11.84SLPEE165 pKa = 4.65GGDD168 pKa = 3.09RR169 pKa = 11.84HH170 pKa = 6.35EE171 pKa = 5.51IEE173 pKa = 5.14ALANHH178 pKa = 6.9KK179 pKa = 10.19AALSSSFEE187 pKa = 4.0VPADD191 pKa = 3.41VLTSLRR197 pKa = 11.84NYY199 pKa = 10.32SEE201 pKa = 3.9DD202 pKa = 2.94WARR205 pKa = 11.84RR206 pKa = 11.84HH207 pKa = 5.84LAADD211 pKa = 4.29PDD213 pKa = 4.17PSLLCEE219 pKa = 4.21PCTGNSATFEE229 pKa = 3.97RR230 pKa = 11.84TRR232 pKa = 11.84RR233 pKa = 11.84EE234 pKa = 3.72GGFAQSITDD243 pKa = 4.3LVSSSPTDD251 pKa = 3.22NLPPLEE257 pKa = 4.51SMPFGPTQGQALPVHH272 pKa = 5.65VLEE275 pKa = 4.8VSLSRR280 pKa = 11.84YY281 pKa = 9.9HH282 pKa = 6.86NGSDD286 pKa = 3.14PKK288 pKa = 11.05GRR290 pKa = 11.84VSVVRR295 pKa = 11.84EE296 pKa = 3.79RR297 pKa = 11.84GHH299 pKa = 5.6KK300 pKa = 9.68VRR302 pKa = 11.84VVSAMEE308 pKa = 3.77THH310 pKa = 6.22EE311 pKa = 4.8LVLGHH316 pKa = 6.83AARR319 pKa = 11.84RR320 pKa = 11.84RR321 pKa = 11.84LFKK324 pKa = 10.41GLRR327 pKa = 11.84RR328 pKa = 11.84EE329 pKa = 3.71RR330 pKa = 11.84RR331 pKa = 11.84LRR333 pKa = 11.84DD334 pKa = 3.27TLKK337 pKa = 11.1GDD339 pKa = 4.37FEE341 pKa = 4.7ATTKK345 pKa = 11.03AFVGCAGTVISSDD358 pKa = 3.26MKK360 pKa = 10.69SASDD364 pKa = 4.96LIPLSVASAIVDD376 pKa = 3.44GLEE379 pKa = 3.82ASGRR383 pKa = 11.84LLPVEE388 pKa = 4.01IAGLRR393 pKa = 11.84ACTGPQHH400 pKa = 6.61LVYY403 pKa = 10.08PDD405 pKa = 3.39GSEE408 pKa = 3.42ITTRR412 pKa = 11.84RR413 pKa = 11.84GILMGLPTTWAILNLMHH430 pKa = 6.89LWCWDD435 pKa = 3.44SADD438 pKa = 3.63RR439 pKa = 11.84QYY441 pKa = 11.59RR442 pKa = 11.84LEE444 pKa = 4.0GHH446 pKa = 6.86PFRR449 pKa = 11.84ATVRR453 pKa = 11.84SDD455 pKa = 3.28CRR457 pKa = 11.84VCGDD461 pKa = 3.98DD462 pKa = 5.4LIGVGPDD469 pKa = 3.06SLLRR473 pKa = 11.84SYY475 pKa = 11.16DD476 pKa = 3.53RR477 pKa = 11.84NLGLVGMILSPGKK490 pKa = 9.72HH491 pKa = 4.72FRR493 pKa = 11.84SNRR496 pKa = 11.84RR497 pKa = 11.84GVFLEE502 pKa = 4.04RR503 pKa = 11.84LLEE506 pKa = 4.08FQTRR510 pKa = 11.84KK511 pKa = 7.36TVYY514 pKa = 8.62EE515 pKa = 3.73HH516 pKa = 6.5AVIYY520 pKa = 10.3RR521 pKa = 11.84KK522 pKa = 9.78VGHH525 pKa = 6.44RR526 pKa = 11.84RR527 pKa = 11.84VPVDD531 pKa = 3.36RR532 pKa = 11.84SHH534 pKa = 7.54IPVVTRR540 pKa = 11.84VTVLNTIPLKK550 pKa = 10.27GLVRR554 pKa = 11.84ASVLGRR560 pKa = 11.84DD561 pKa = 4.72DD562 pKa = 4.49PPVWWAAAVAEE573 pKa = 4.35SSLLSDD579 pKa = 3.85YY580 pKa = 10.39PRR582 pKa = 11.84KK583 pKa = 10.3KK584 pKa = 9.78IFAAARR590 pKa = 11.84TLRR593 pKa = 11.84PGLSRR598 pKa = 11.84QFRR601 pKa = 11.84RR602 pKa = 11.84LGIPPFLPRR611 pKa = 11.84EE612 pKa = 3.97LGGAGLVGPSDD623 pKa = 4.21RR624 pKa = 11.84VDD626 pKa = 3.23APAFHH631 pKa = 7.13RR632 pKa = 11.84KK633 pKa = 9.49AISSLVWGSDD643 pKa = 2.48ATAAYY648 pKa = 10.48SFIRR652 pKa = 11.84MWQGFEE658 pKa = 3.63GHH660 pKa = 6.32PWKK663 pKa = 9.11TAASQEE669 pKa = 4.06TDD671 pKa = 2.49TWFADD676 pKa = 3.76YY677 pKa = 10.92KK678 pKa = 8.56VTRR681 pKa = 11.84PGKK684 pKa = 9.43MYY686 pKa = 9.43PDD688 pKa = 3.43RR689 pKa = 11.84YY690 pKa = 10.6GFLDD694 pKa = 4.04GEE696 pKa = 4.45SLRR699 pKa = 11.84TKK701 pKa = 9.98STMLNSAVYY710 pKa = 8.14EE711 pKa = 4.36TFLGPDD717 pKa = 3.94PDD719 pKa = 3.78ATHH722 pKa = 6.23YY723 pKa = 9.92PSLRR727 pKa = 11.84IVASRR732 pKa = 11.84LAKK735 pKa = 9.74VRR737 pKa = 11.84KK738 pKa = 9.62DD739 pKa = 3.49LVNRR743 pKa = 11.84WPSVKK748 pKa = 10.06PVGKK752 pKa = 10.04DD753 pKa = 2.69LGTILEE759 pKa = 4.41AFEE762 pKa = 4.07EE763 pKa = 4.68SKK765 pKa = 11.25LCTLWVTPYY774 pKa = 10.49DD775 pKa = 3.46ASGYY779 pKa = 10.41FDD781 pKa = 6.02DD782 pKa = 5.96SLLLMDD788 pKa = 4.15EE789 pKa = 4.39SVYY792 pKa = 10.06QRR794 pKa = 11.84RR795 pKa = 11.84FRR797 pKa = 11.84QLVIAGLMRR806 pKa = 11.84EE807 pKa = 4.13GRR809 pKa = 11.84MGDD812 pKa = 3.56LLFPNWLPPSTVVSGFPP829 pKa = 3.46
MM1 pKa = 7.68KK2 pKa = 10.46EE3 pKa = 3.87PVDD6 pKa = 4.16CRR8 pKa = 11.84LSTPAGFSGTVPPPGRR24 pKa = 11.84TKK26 pKa = 10.44AARR29 pKa = 11.84PGTIPVRR36 pKa = 11.84RR37 pKa = 11.84SRR39 pKa = 11.84GSASALPGKK48 pKa = 10.24IYY50 pKa = 10.22GWSRR54 pKa = 11.84RR55 pKa = 11.84QRR57 pKa = 11.84DD58 pKa = 3.69RR59 pKa = 11.84FAMLLSSFDD68 pKa = 3.67AALAAYY74 pKa = 9.68SGVVVSRR81 pKa = 11.84GTRR84 pKa = 11.84SLPPSLRR91 pKa = 11.84LFRR94 pKa = 11.84AMTRR98 pKa = 11.84KK99 pKa = 8.74WLSVTARR106 pKa = 11.84GNGVEE111 pKa = 4.18FAIASAKK118 pKa = 9.88EE119 pKa = 3.6FSAACRR125 pKa = 11.84AGWISGTVPDD135 pKa = 4.5HH136 pKa = 7.26FFMKK140 pKa = 10.09WLPEE144 pKa = 3.67PVRR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 10.1SGLWAQLSFIGRR161 pKa = 11.84SLPEE165 pKa = 4.65GGDD168 pKa = 3.09RR169 pKa = 11.84HH170 pKa = 6.35EE171 pKa = 5.51IEE173 pKa = 5.14ALANHH178 pKa = 6.9KK179 pKa = 10.19AALSSSFEE187 pKa = 4.0VPADD191 pKa = 3.41VLTSLRR197 pKa = 11.84NYY199 pKa = 10.32SEE201 pKa = 3.9DD202 pKa = 2.94WARR205 pKa = 11.84RR206 pKa = 11.84HH207 pKa = 5.84LAADD211 pKa = 4.29PDD213 pKa = 4.17PSLLCEE219 pKa = 4.21PCTGNSATFEE229 pKa = 3.97RR230 pKa = 11.84TRR232 pKa = 11.84RR233 pKa = 11.84EE234 pKa = 3.72GGFAQSITDD243 pKa = 4.3LVSSSPTDD251 pKa = 3.22NLPPLEE257 pKa = 4.51SMPFGPTQGQALPVHH272 pKa = 5.65VLEE275 pKa = 4.8VSLSRR280 pKa = 11.84YY281 pKa = 9.9HH282 pKa = 6.86NGSDD286 pKa = 3.14PKK288 pKa = 11.05GRR290 pKa = 11.84VSVVRR295 pKa = 11.84EE296 pKa = 3.79RR297 pKa = 11.84GHH299 pKa = 5.6KK300 pKa = 9.68VRR302 pKa = 11.84VVSAMEE308 pKa = 3.77THH310 pKa = 6.22EE311 pKa = 4.8LVLGHH316 pKa = 6.83AARR319 pKa = 11.84RR320 pKa = 11.84RR321 pKa = 11.84LFKK324 pKa = 10.41GLRR327 pKa = 11.84RR328 pKa = 11.84EE329 pKa = 3.71RR330 pKa = 11.84RR331 pKa = 11.84LRR333 pKa = 11.84DD334 pKa = 3.27TLKK337 pKa = 11.1GDD339 pKa = 4.37FEE341 pKa = 4.7ATTKK345 pKa = 11.03AFVGCAGTVISSDD358 pKa = 3.26MKK360 pKa = 10.69SASDD364 pKa = 4.96LIPLSVASAIVDD376 pKa = 3.44GLEE379 pKa = 3.82ASGRR383 pKa = 11.84LLPVEE388 pKa = 4.01IAGLRR393 pKa = 11.84ACTGPQHH400 pKa = 6.61LVYY403 pKa = 10.08PDD405 pKa = 3.39GSEE408 pKa = 3.42ITTRR412 pKa = 11.84RR413 pKa = 11.84GILMGLPTTWAILNLMHH430 pKa = 6.89LWCWDD435 pKa = 3.44SADD438 pKa = 3.63RR439 pKa = 11.84QYY441 pKa = 11.59RR442 pKa = 11.84LEE444 pKa = 4.0GHH446 pKa = 6.86PFRR449 pKa = 11.84ATVRR453 pKa = 11.84SDD455 pKa = 3.28CRR457 pKa = 11.84VCGDD461 pKa = 3.98DD462 pKa = 5.4LIGVGPDD469 pKa = 3.06SLLRR473 pKa = 11.84SYY475 pKa = 11.16DD476 pKa = 3.53RR477 pKa = 11.84NLGLVGMILSPGKK490 pKa = 9.72HH491 pKa = 4.72FRR493 pKa = 11.84SNRR496 pKa = 11.84RR497 pKa = 11.84GVFLEE502 pKa = 4.04RR503 pKa = 11.84LLEE506 pKa = 4.08FQTRR510 pKa = 11.84KK511 pKa = 7.36TVYY514 pKa = 8.62EE515 pKa = 3.73HH516 pKa = 6.5AVIYY520 pKa = 10.3RR521 pKa = 11.84KK522 pKa = 9.78VGHH525 pKa = 6.44RR526 pKa = 11.84RR527 pKa = 11.84VPVDD531 pKa = 3.36RR532 pKa = 11.84SHH534 pKa = 7.54IPVVTRR540 pKa = 11.84VTVLNTIPLKK550 pKa = 10.27GLVRR554 pKa = 11.84ASVLGRR560 pKa = 11.84DD561 pKa = 4.72DD562 pKa = 4.49PPVWWAAAVAEE573 pKa = 4.35SSLLSDD579 pKa = 3.85YY580 pKa = 10.39PRR582 pKa = 11.84KK583 pKa = 10.3KK584 pKa = 9.78IFAAARR590 pKa = 11.84TLRR593 pKa = 11.84PGLSRR598 pKa = 11.84QFRR601 pKa = 11.84RR602 pKa = 11.84LGIPPFLPRR611 pKa = 11.84EE612 pKa = 3.97LGGAGLVGPSDD623 pKa = 4.21RR624 pKa = 11.84VDD626 pKa = 3.23APAFHH631 pKa = 7.13RR632 pKa = 11.84KK633 pKa = 9.49AISSLVWGSDD643 pKa = 2.48ATAAYY648 pKa = 10.48SFIRR652 pKa = 11.84MWQGFEE658 pKa = 3.63GHH660 pKa = 6.32PWKK663 pKa = 9.11TAASQEE669 pKa = 4.06TDD671 pKa = 2.49TWFADD676 pKa = 3.76YY677 pKa = 10.92KK678 pKa = 8.56VTRR681 pKa = 11.84PGKK684 pKa = 9.43MYY686 pKa = 9.43PDD688 pKa = 3.43RR689 pKa = 11.84YY690 pKa = 10.6GFLDD694 pKa = 4.04GEE696 pKa = 4.45SLRR699 pKa = 11.84TKK701 pKa = 9.98STMLNSAVYY710 pKa = 8.14EE711 pKa = 4.36TFLGPDD717 pKa = 3.94PDD719 pKa = 3.78ATHH722 pKa = 6.23YY723 pKa = 9.92PSLRR727 pKa = 11.84IVASRR732 pKa = 11.84LAKK735 pKa = 9.74VRR737 pKa = 11.84KK738 pKa = 9.62DD739 pKa = 3.49LVNRR743 pKa = 11.84WPSVKK748 pKa = 10.06PVGKK752 pKa = 10.04DD753 pKa = 2.69LGTILEE759 pKa = 4.41AFEE762 pKa = 4.07EE763 pKa = 4.68SKK765 pKa = 11.25LCTLWVTPYY774 pKa = 10.49DD775 pKa = 3.46ASGYY779 pKa = 10.41FDD781 pKa = 6.02DD782 pKa = 5.96SLLLMDD788 pKa = 4.15EE789 pKa = 4.39SVYY792 pKa = 10.06QRR794 pKa = 11.84RR795 pKa = 11.84FRR797 pKa = 11.84QLVIAGLMRR806 pKa = 11.84EE807 pKa = 4.13GRR809 pKa = 11.84MGDD812 pKa = 3.56LLFPNWLPPSTVVSGFPP829 pKa = 3.46
Molecular weight: 91.81 kDa
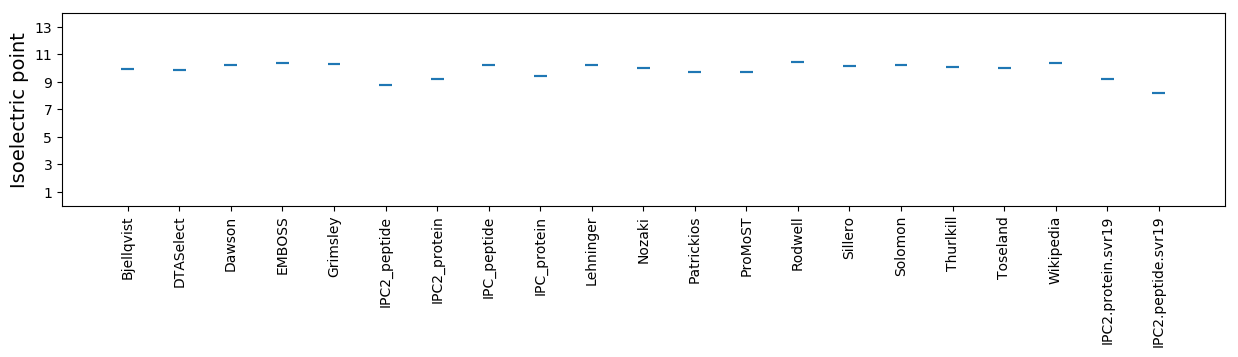
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P25328|RDRP_SCV20 RNA-directed RNA polymerase OS=Saccharomyces 20S RNA narnavirus OX=186772 PE=1 SV=1
MM1 pKa = 7.68KK2 pKa = 10.46EE3 pKa = 3.87PVDD6 pKa = 4.16CRR8 pKa = 11.84LSTPAGFSGTVPPPGRR24 pKa = 11.84TKK26 pKa = 10.44AARR29 pKa = 11.84PGTIPVRR36 pKa = 11.84RR37 pKa = 11.84SRR39 pKa = 11.84GSASALPGKK48 pKa = 10.24IYY50 pKa = 10.22GWSRR54 pKa = 11.84RR55 pKa = 11.84QRR57 pKa = 11.84DD58 pKa = 3.69RR59 pKa = 11.84FAMLLSSFDD68 pKa = 3.67AALAAYY74 pKa = 9.68SGVVVSRR81 pKa = 11.84GTRR84 pKa = 11.84SLPPSLRR91 pKa = 11.84LFRR94 pKa = 11.84AMTRR98 pKa = 11.84KK99 pKa = 8.74WLSVTARR106 pKa = 11.84GNGVEE111 pKa = 4.18FAIASAKK118 pKa = 9.88EE119 pKa = 3.6FSAACRR125 pKa = 11.84AGWISGTVPDD135 pKa = 4.5HH136 pKa = 7.26FFMKK140 pKa = 10.09WLPEE144 pKa = 3.67PVRR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 10.1SGLWAQLSFIGRR161 pKa = 11.84SLPEE165 pKa = 4.65GGDD168 pKa = 3.09RR169 pKa = 11.84HH170 pKa = 6.35EE171 pKa = 5.51IEE173 pKa = 5.14ALANHH178 pKa = 6.9KK179 pKa = 10.19AALSSSFEE187 pKa = 4.0VPADD191 pKa = 3.41VLTSLRR197 pKa = 11.84NYY199 pKa = 10.32SEE201 pKa = 3.9DD202 pKa = 2.94WARR205 pKa = 11.84RR206 pKa = 11.84HH207 pKa = 5.84LAADD211 pKa = 4.29PDD213 pKa = 4.17PSLLCEE219 pKa = 4.21PCTGNSATFEE229 pKa = 3.97RR230 pKa = 11.84TRR232 pKa = 11.84RR233 pKa = 11.84EE234 pKa = 3.72GGFAQSITDD243 pKa = 4.3LVSSSPTDD251 pKa = 3.22NLPPLEE257 pKa = 4.51SMPFGPTQGQALPVHH272 pKa = 5.65VLEE275 pKa = 4.8VSLSRR280 pKa = 11.84YY281 pKa = 9.9HH282 pKa = 6.86NGSDD286 pKa = 3.14PKK288 pKa = 11.05GRR290 pKa = 11.84VSVVRR295 pKa = 11.84EE296 pKa = 3.79RR297 pKa = 11.84GHH299 pKa = 5.6KK300 pKa = 9.68VRR302 pKa = 11.84VVSAMEE308 pKa = 3.77THH310 pKa = 6.22EE311 pKa = 4.8LVLGHH316 pKa = 6.83AARR319 pKa = 11.84RR320 pKa = 11.84RR321 pKa = 11.84LFKK324 pKa = 10.41GLRR327 pKa = 11.84RR328 pKa = 11.84EE329 pKa = 3.71RR330 pKa = 11.84RR331 pKa = 11.84LRR333 pKa = 11.84DD334 pKa = 3.27TLKK337 pKa = 11.1GDD339 pKa = 4.37FEE341 pKa = 4.7ATTKK345 pKa = 11.03AFVGCAGTVISSDD358 pKa = 3.26MKK360 pKa = 10.69SASDD364 pKa = 4.96LIPLSVASAIVDD376 pKa = 3.44GLEE379 pKa = 3.82ASGRR383 pKa = 11.84LLPVEE388 pKa = 4.01IAGLRR393 pKa = 11.84ACTGPQHH400 pKa = 6.61LVYY403 pKa = 10.08PDD405 pKa = 3.39GSEE408 pKa = 3.42ITTRR412 pKa = 11.84RR413 pKa = 11.84GILMGLPTTWAILNLMHH430 pKa = 6.89LWCWDD435 pKa = 3.44SADD438 pKa = 3.63RR439 pKa = 11.84QYY441 pKa = 11.59RR442 pKa = 11.84LEE444 pKa = 4.0GHH446 pKa = 6.86PFRR449 pKa = 11.84ATVRR453 pKa = 11.84SDD455 pKa = 3.28CRR457 pKa = 11.84VCGDD461 pKa = 3.98DD462 pKa = 5.4LIGVGPDD469 pKa = 3.06SLLRR473 pKa = 11.84SYY475 pKa = 11.16DD476 pKa = 3.53RR477 pKa = 11.84NLGLVGMILSPGKK490 pKa = 9.72HH491 pKa = 4.72FRR493 pKa = 11.84SNRR496 pKa = 11.84RR497 pKa = 11.84GVFLEE502 pKa = 4.04RR503 pKa = 11.84LLEE506 pKa = 4.08FQTRR510 pKa = 11.84KK511 pKa = 7.36TVYY514 pKa = 8.62EE515 pKa = 3.73HH516 pKa = 6.5AVIYY520 pKa = 10.3RR521 pKa = 11.84KK522 pKa = 9.78VGHH525 pKa = 6.44RR526 pKa = 11.84RR527 pKa = 11.84VPVDD531 pKa = 3.36RR532 pKa = 11.84SHH534 pKa = 7.54IPVVTRR540 pKa = 11.84VTVLNTIPLKK550 pKa = 10.27GLVRR554 pKa = 11.84ASVLGRR560 pKa = 11.84DD561 pKa = 4.72DD562 pKa = 4.49PPVWWAAAVAEE573 pKa = 4.35SSLLSDD579 pKa = 3.85YY580 pKa = 10.39PRR582 pKa = 11.84KK583 pKa = 10.3KK584 pKa = 9.78IFAAARR590 pKa = 11.84TLRR593 pKa = 11.84PGLSRR598 pKa = 11.84QFRR601 pKa = 11.84RR602 pKa = 11.84LGIPPFLPRR611 pKa = 11.84EE612 pKa = 3.97LGGAGLVGPSDD623 pKa = 4.21RR624 pKa = 11.84VDD626 pKa = 3.23APAFHH631 pKa = 7.13RR632 pKa = 11.84KK633 pKa = 9.49AISSLVWGSDD643 pKa = 2.48ATAAYY648 pKa = 10.48SFIRR652 pKa = 11.84MWQGFEE658 pKa = 3.63GHH660 pKa = 6.32PWKK663 pKa = 9.11TAASQEE669 pKa = 4.06TDD671 pKa = 2.49TWFADD676 pKa = 3.76YY677 pKa = 10.92KK678 pKa = 8.56VTRR681 pKa = 11.84PGKK684 pKa = 9.43MYY686 pKa = 9.43PDD688 pKa = 3.43RR689 pKa = 11.84YY690 pKa = 10.6GFLDD694 pKa = 4.04GEE696 pKa = 4.45SLRR699 pKa = 11.84TKK701 pKa = 9.98STMLNSAVYY710 pKa = 8.14EE711 pKa = 4.36TFLGPDD717 pKa = 3.94PDD719 pKa = 3.78ATHH722 pKa = 6.23YY723 pKa = 9.92PSLRR727 pKa = 11.84IVASRR732 pKa = 11.84LAKK735 pKa = 9.74VRR737 pKa = 11.84KK738 pKa = 9.62DD739 pKa = 3.49LVNRR743 pKa = 11.84WPSVKK748 pKa = 10.06PVGKK752 pKa = 10.04DD753 pKa = 2.69LGTILEE759 pKa = 4.41AFEE762 pKa = 4.07EE763 pKa = 4.68SKK765 pKa = 11.25LCTLWVTPYY774 pKa = 10.49DD775 pKa = 3.46ASGYY779 pKa = 10.41FDD781 pKa = 6.02DD782 pKa = 5.96SLLLMDD788 pKa = 4.15EE789 pKa = 4.39SVYY792 pKa = 10.06QRR794 pKa = 11.84RR795 pKa = 11.84FRR797 pKa = 11.84QLVIAGLMRR806 pKa = 11.84EE807 pKa = 4.13GRR809 pKa = 11.84MGDD812 pKa = 3.56LLFPNWLPPSTVVSGFPP829 pKa = 3.46
MM1 pKa = 7.68KK2 pKa = 10.46EE3 pKa = 3.87PVDD6 pKa = 4.16CRR8 pKa = 11.84LSTPAGFSGTVPPPGRR24 pKa = 11.84TKK26 pKa = 10.44AARR29 pKa = 11.84PGTIPVRR36 pKa = 11.84RR37 pKa = 11.84SRR39 pKa = 11.84GSASALPGKK48 pKa = 10.24IYY50 pKa = 10.22GWSRR54 pKa = 11.84RR55 pKa = 11.84QRR57 pKa = 11.84DD58 pKa = 3.69RR59 pKa = 11.84FAMLLSSFDD68 pKa = 3.67AALAAYY74 pKa = 9.68SGVVVSRR81 pKa = 11.84GTRR84 pKa = 11.84SLPPSLRR91 pKa = 11.84LFRR94 pKa = 11.84AMTRR98 pKa = 11.84KK99 pKa = 8.74WLSVTARR106 pKa = 11.84GNGVEE111 pKa = 4.18FAIASAKK118 pKa = 9.88EE119 pKa = 3.6FSAACRR125 pKa = 11.84AGWISGTVPDD135 pKa = 4.5HH136 pKa = 7.26FFMKK140 pKa = 10.09WLPEE144 pKa = 3.67PVRR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 10.1SGLWAQLSFIGRR161 pKa = 11.84SLPEE165 pKa = 4.65GGDD168 pKa = 3.09RR169 pKa = 11.84HH170 pKa = 6.35EE171 pKa = 5.51IEE173 pKa = 5.14ALANHH178 pKa = 6.9KK179 pKa = 10.19AALSSSFEE187 pKa = 4.0VPADD191 pKa = 3.41VLTSLRR197 pKa = 11.84NYY199 pKa = 10.32SEE201 pKa = 3.9DD202 pKa = 2.94WARR205 pKa = 11.84RR206 pKa = 11.84HH207 pKa = 5.84LAADD211 pKa = 4.29PDD213 pKa = 4.17PSLLCEE219 pKa = 4.21PCTGNSATFEE229 pKa = 3.97RR230 pKa = 11.84TRR232 pKa = 11.84RR233 pKa = 11.84EE234 pKa = 3.72GGFAQSITDD243 pKa = 4.3LVSSSPTDD251 pKa = 3.22NLPPLEE257 pKa = 4.51SMPFGPTQGQALPVHH272 pKa = 5.65VLEE275 pKa = 4.8VSLSRR280 pKa = 11.84YY281 pKa = 9.9HH282 pKa = 6.86NGSDD286 pKa = 3.14PKK288 pKa = 11.05GRR290 pKa = 11.84VSVVRR295 pKa = 11.84EE296 pKa = 3.79RR297 pKa = 11.84GHH299 pKa = 5.6KK300 pKa = 9.68VRR302 pKa = 11.84VVSAMEE308 pKa = 3.77THH310 pKa = 6.22EE311 pKa = 4.8LVLGHH316 pKa = 6.83AARR319 pKa = 11.84RR320 pKa = 11.84RR321 pKa = 11.84LFKK324 pKa = 10.41GLRR327 pKa = 11.84RR328 pKa = 11.84EE329 pKa = 3.71RR330 pKa = 11.84RR331 pKa = 11.84LRR333 pKa = 11.84DD334 pKa = 3.27TLKK337 pKa = 11.1GDD339 pKa = 4.37FEE341 pKa = 4.7ATTKK345 pKa = 11.03AFVGCAGTVISSDD358 pKa = 3.26MKK360 pKa = 10.69SASDD364 pKa = 4.96LIPLSVASAIVDD376 pKa = 3.44GLEE379 pKa = 3.82ASGRR383 pKa = 11.84LLPVEE388 pKa = 4.01IAGLRR393 pKa = 11.84ACTGPQHH400 pKa = 6.61LVYY403 pKa = 10.08PDD405 pKa = 3.39GSEE408 pKa = 3.42ITTRR412 pKa = 11.84RR413 pKa = 11.84GILMGLPTTWAILNLMHH430 pKa = 6.89LWCWDD435 pKa = 3.44SADD438 pKa = 3.63RR439 pKa = 11.84QYY441 pKa = 11.59RR442 pKa = 11.84LEE444 pKa = 4.0GHH446 pKa = 6.86PFRR449 pKa = 11.84ATVRR453 pKa = 11.84SDD455 pKa = 3.28CRR457 pKa = 11.84VCGDD461 pKa = 3.98DD462 pKa = 5.4LIGVGPDD469 pKa = 3.06SLLRR473 pKa = 11.84SYY475 pKa = 11.16DD476 pKa = 3.53RR477 pKa = 11.84NLGLVGMILSPGKK490 pKa = 9.72HH491 pKa = 4.72FRR493 pKa = 11.84SNRR496 pKa = 11.84RR497 pKa = 11.84GVFLEE502 pKa = 4.04RR503 pKa = 11.84LLEE506 pKa = 4.08FQTRR510 pKa = 11.84KK511 pKa = 7.36TVYY514 pKa = 8.62EE515 pKa = 3.73HH516 pKa = 6.5AVIYY520 pKa = 10.3RR521 pKa = 11.84KK522 pKa = 9.78VGHH525 pKa = 6.44RR526 pKa = 11.84RR527 pKa = 11.84VPVDD531 pKa = 3.36RR532 pKa = 11.84SHH534 pKa = 7.54IPVVTRR540 pKa = 11.84VTVLNTIPLKK550 pKa = 10.27GLVRR554 pKa = 11.84ASVLGRR560 pKa = 11.84DD561 pKa = 4.72DD562 pKa = 4.49PPVWWAAAVAEE573 pKa = 4.35SSLLSDD579 pKa = 3.85YY580 pKa = 10.39PRR582 pKa = 11.84KK583 pKa = 10.3KK584 pKa = 9.78IFAAARR590 pKa = 11.84TLRR593 pKa = 11.84PGLSRR598 pKa = 11.84QFRR601 pKa = 11.84RR602 pKa = 11.84LGIPPFLPRR611 pKa = 11.84EE612 pKa = 3.97LGGAGLVGPSDD623 pKa = 4.21RR624 pKa = 11.84VDD626 pKa = 3.23APAFHH631 pKa = 7.13RR632 pKa = 11.84KK633 pKa = 9.49AISSLVWGSDD643 pKa = 2.48ATAAYY648 pKa = 10.48SFIRR652 pKa = 11.84MWQGFEE658 pKa = 3.63GHH660 pKa = 6.32PWKK663 pKa = 9.11TAASQEE669 pKa = 4.06TDD671 pKa = 2.49TWFADD676 pKa = 3.76YY677 pKa = 10.92KK678 pKa = 8.56VTRR681 pKa = 11.84PGKK684 pKa = 9.43MYY686 pKa = 9.43PDD688 pKa = 3.43RR689 pKa = 11.84YY690 pKa = 10.6GFLDD694 pKa = 4.04GEE696 pKa = 4.45SLRR699 pKa = 11.84TKK701 pKa = 9.98STMLNSAVYY710 pKa = 8.14EE711 pKa = 4.36TFLGPDD717 pKa = 3.94PDD719 pKa = 3.78ATHH722 pKa = 6.23YY723 pKa = 9.92PSLRR727 pKa = 11.84IVASRR732 pKa = 11.84LAKK735 pKa = 9.74VRR737 pKa = 11.84KK738 pKa = 9.62DD739 pKa = 3.49LVNRR743 pKa = 11.84WPSVKK748 pKa = 10.06PVGKK752 pKa = 10.04DD753 pKa = 2.69LGTILEE759 pKa = 4.41AFEE762 pKa = 4.07EE763 pKa = 4.68SKK765 pKa = 11.25LCTLWVTPYY774 pKa = 10.49DD775 pKa = 3.46ASGYY779 pKa = 10.41FDD781 pKa = 6.02DD782 pKa = 5.96SLLLMDD788 pKa = 4.15EE789 pKa = 4.39SVYY792 pKa = 10.06QRR794 pKa = 11.84RR795 pKa = 11.84FRR797 pKa = 11.84QLVIAGLMRR806 pKa = 11.84EE807 pKa = 4.13GRR809 pKa = 11.84MGDD812 pKa = 3.56LLFPNWLPPSTVVSGFPP829 pKa = 3.46
Molecular weight: 91.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
829 |
829 |
829 |
829.0 |
91.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.565 ± 0.0 | 1.206 ± 0.0 |
5.428 ± 0.0 | 4.463 ± 0.0 |
4.101 ± 0.0 | 8.203 ± 0.0 |
2.292 ± 0.0 | 3.136 ± 0.0 |
3.619 ± 0.0 | 10.374 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.93 ± 0.0 | 1.568 ± 0.0 |
6.996 ± 0.0 | 1.568 ± 0.0 |
10.253 ± 0.0 | 8.926 ± 0.0 |
5.428 ± 0.0 | 7.479 ± 0.0 |
2.171 ± 0.0 | 2.292 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
