
Vibrio phage VP5
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Enhodamvirus; Vibrio virus VP5
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
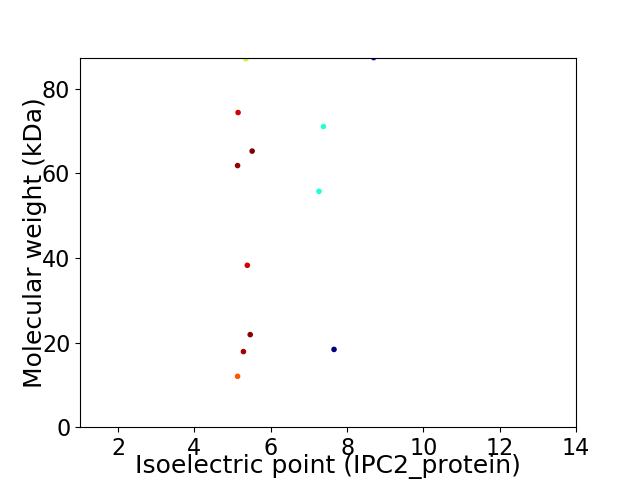
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
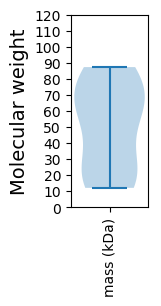
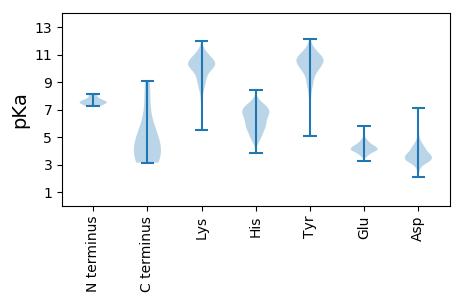
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q6R6B3|Q6R6B3_9CAUD Peptidase_M15_3 domain-containing protein OS=Vibrio phage VP5 OX=260827 PE=4 SV=1
MM1 pKa = 7.66PSLVSIYY8 pKa = 10.77NKK10 pKa = 10.52ALAHH14 pKa = 5.83VGAKK18 pKa = 9.58PVLSEE23 pKa = 3.67TDD25 pKa = 3.1ANTRR29 pKa = 11.84AEE31 pKa = 4.37MCNAMYY37 pKa = 10.65EE38 pKa = 4.25FALRR42 pKa = 11.84SALEE46 pKa = 3.75ARR48 pKa = 11.84EE49 pKa = 3.86WSFAAMRR56 pKa = 11.84VRR58 pKa = 11.84LLPDD62 pKa = 3.81PLITPAWGYY71 pKa = 10.66NYY73 pKa = 10.45GYY75 pKa = 10.44RR76 pKa = 11.84LPNDD80 pKa = 4.68CIRR83 pKa = 11.84VCEE86 pKa = 4.05VRR88 pKa = 11.84DD89 pKa = 3.81GNSYY93 pKa = 10.75NIDD96 pKa = 3.1WAIEE100 pKa = 4.14SGFLYY105 pKa = 9.28ATRR108 pKa = 11.84PTVDD112 pKa = 2.62VKK114 pKa = 10.86YY115 pKa = 10.83NRR117 pKa = 11.84LVEE120 pKa = 4.45DD121 pKa = 4.45PNAFTPTFVEE131 pKa = 4.11ALSLKK136 pKa = 9.98LASDD140 pKa = 3.57ICIPLTEE147 pKa = 4.13NRR149 pKa = 11.84GLGNDD154 pKa = 4.46LISRR158 pKa = 11.84YY159 pKa = 10.29LMAVSEE165 pKa = 4.4GAALDD170 pKa = 3.69GLQASRR176 pKa = 11.84EE177 pKa = 3.98RR178 pKa = 11.84LRR180 pKa = 11.84ADD182 pKa = 3.3TLINARR188 pKa = 11.84RR189 pKa = 11.84GCGRR193 pKa = 11.84MDD195 pKa = 3.44RR196 pKa = 11.84PYY198 pKa = 10.28PQTGSGKK205 pKa = 10.13FYY207 pKa = 10.65DD208 pKa = 4.03IAPPTIPDD216 pKa = 3.74SPPVGNISGPSSVQLGQPFSFIAQVTGTQPISYY249 pKa = 8.76QWYY252 pKa = 8.64KK253 pKa = 11.09NGIAIPSATSVSFGVEE269 pKa = 3.66SATNNDD275 pKa = 3.14EE276 pKa = 4.88GEE278 pKa = 4.34YY279 pKa = 10.46HH280 pKa = 7.03CLFSNSAGVVISNKK294 pKa = 9.05ISLEE298 pKa = 3.94IGCFPPVHH306 pKa = 6.77NITGPTTVGEE316 pKa = 4.37GKK318 pKa = 10.04SFSMVCTVTAGNLPVTYY335 pKa = 9.76QWQRR339 pKa = 11.84DD340 pKa = 3.73GQNIAGATTATLSVAAATVVDD361 pKa = 3.99SGEE364 pKa = 4.04YY365 pKa = 10.29DD366 pKa = 3.42CLVSNQCGSNVSSTNKK382 pKa = 8.22ITLDD386 pKa = 3.79VIPLVVPTVSISPVSASPLEE406 pKa = 4.13PAPVTFTASVVDD418 pKa = 4.82DD419 pKa = 4.5GGAPPVTLKK428 pKa = 10.32WYY430 pKa = 10.86LNGNLVQNGGTTYY443 pKa = 9.97TSPPTAVGQNRR454 pKa = 11.84TVSVVGTNSVGDD466 pKa = 4.15SVPASASLTPVSNFISQSTFTTNAAFTLHH495 pKa = 7.18PDD497 pKa = 3.38CTFVQVQGCGGGGGGGSGDD516 pKa = 4.23AIGTEE521 pKa = 4.25LTGGGGGGGAALVSVLSSNAIGGQTATIVIGGGGAGAPNVGTTFGGNGGNTSVSGAGIIGLSWTGGKK588 pKa = 9.82GGRR591 pKa = 11.84SGYY594 pKa = 10.43RR595 pKa = 11.84SDD597 pKa = 4.04GSGNGAFGGAGGNNSANTGGGAGGVSSNGGNGVNGGGGGGGSNNDD642 pKa = 2.96LVLRR646 pKa = 11.84RR647 pKa = 11.84GGNGGGSNGGIGGQGVGTTDD667 pKa = 3.51GPSGGGGGNSAVWWDD682 pKa = 3.45NADD685 pKa = 3.43AGRR688 pKa = 11.84GGNGVTGNSGNQTGGHH704 pKa = 5.5ATGNGNGGGGGGGGVPDD721 pKa = 4.59FGGAGGNGSGGKK733 pKa = 10.12VIIRR737 pKa = 11.84QYY739 pKa = 10.65RR740 pKa = 11.84RR741 pKa = 11.84PII743 pKa = 3.5
MM1 pKa = 7.66PSLVSIYY8 pKa = 10.77NKK10 pKa = 10.52ALAHH14 pKa = 5.83VGAKK18 pKa = 9.58PVLSEE23 pKa = 3.67TDD25 pKa = 3.1ANTRR29 pKa = 11.84AEE31 pKa = 4.37MCNAMYY37 pKa = 10.65EE38 pKa = 4.25FALRR42 pKa = 11.84SALEE46 pKa = 3.75ARR48 pKa = 11.84EE49 pKa = 3.86WSFAAMRR56 pKa = 11.84VRR58 pKa = 11.84LLPDD62 pKa = 3.81PLITPAWGYY71 pKa = 10.66NYY73 pKa = 10.45GYY75 pKa = 10.44RR76 pKa = 11.84LPNDD80 pKa = 4.68CIRR83 pKa = 11.84VCEE86 pKa = 4.05VRR88 pKa = 11.84DD89 pKa = 3.81GNSYY93 pKa = 10.75NIDD96 pKa = 3.1WAIEE100 pKa = 4.14SGFLYY105 pKa = 9.28ATRR108 pKa = 11.84PTVDD112 pKa = 2.62VKK114 pKa = 10.86YY115 pKa = 10.83NRR117 pKa = 11.84LVEE120 pKa = 4.45DD121 pKa = 4.45PNAFTPTFVEE131 pKa = 4.11ALSLKK136 pKa = 9.98LASDD140 pKa = 3.57ICIPLTEE147 pKa = 4.13NRR149 pKa = 11.84GLGNDD154 pKa = 4.46LISRR158 pKa = 11.84YY159 pKa = 10.29LMAVSEE165 pKa = 4.4GAALDD170 pKa = 3.69GLQASRR176 pKa = 11.84EE177 pKa = 3.98RR178 pKa = 11.84LRR180 pKa = 11.84ADD182 pKa = 3.3TLINARR188 pKa = 11.84RR189 pKa = 11.84GCGRR193 pKa = 11.84MDD195 pKa = 3.44RR196 pKa = 11.84PYY198 pKa = 10.28PQTGSGKK205 pKa = 10.13FYY207 pKa = 10.65DD208 pKa = 4.03IAPPTIPDD216 pKa = 3.74SPPVGNISGPSSVQLGQPFSFIAQVTGTQPISYY249 pKa = 8.76QWYY252 pKa = 8.64KK253 pKa = 11.09NGIAIPSATSVSFGVEE269 pKa = 3.66SATNNDD275 pKa = 3.14EE276 pKa = 4.88GEE278 pKa = 4.34YY279 pKa = 10.46HH280 pKa = 7.03CLFSNSAGVVISNKK294 pKa = 9.05ISLEE298 pKa = 3.94IGCFPPVHH306 pKa = 6.77NITGPTTVGEE316 pKa = 4.37GKK318 pKa = 10.04SFSMVCTVTAGNLPVTYY335 pKa = 9.76QWQRR339 pKa = 11.84DD340 pKa = 3.73GQNIAGATTATLSVAAATVVDD361 pKa = 3.99SGEE364 pKa = 4.04YY365 pKa = 10.29DD366 pKa = 3.42CLVSNQCGSNVSSTNKK382 pKa = 8.22ITLDD386 pKa = 3.79VIPLVVPTVSISPVSASPLEE406 pKa = 4.13PAPVTFTASVVDD418 pKa = 4.82DD419 pKa = 4.5GGAPPVTLKK428 pKa = 10.32WYY430 pKa = 10.86LNGNLVQNGGTTYY443 pKa = 9.97TSPPTAVGQNRR454 pKa = 11.84TVSVVGTNSVGDD466 pKa = 4.15SVPASASLTPVSNFISQSTFTTNAAFTLHH495 pKa = 7.18PDD497 pKa = 3.38CTFVQVQGCGGGGGGGSGDD516 pKa = 4.23AIGTEE521 pKa = 4.25LTGGGGGGGAALVSVLSSNAIGGQTATIVIGGGGAGAPNVGTTFGGNGGNTSVSGAGIIGLSWTGGKK588 pKa = 9.82GGRR591 pKa = 11.84SGYY594 pKa = 10.43RR595 pKa = 11.84SDD597 pKa = 4.04GSGNGAFGGAGGNNSANTGGGAGGVSSNGGNGVNGGGGGGGSNNDD642 pKa = 2.96LVLRR646 pKa = 11.84RR647 pKa = 11.84GGNGGGSNGGIGGQGVGTTDD667 pKa = 3.51GPSGGGGGNSAVWWDD682 pKa = 3.45NADD685 pKa = 3.43AGRR688 pKa = 11.84GGNGVTGNSGNQTGGHH704 pKa = 5.5ATGNGNGGGGGGGGVPDD721 pKa = 4.59FGGAGGNGSGGKK733 pKa = 10.12VIIRR737 pKa = 11.84QYY739 pKa = 10.65RR740 pKa = 11.84RR741 pKa = 11.84PII743 pKa = 3.5
Molecular weight: 74.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6R6B2|Q6R6B2_9CAUD Major tail subunit OS=Vibrio phage VP5 OX=260827 PE=4 SV=1
MM1 pKa = 7.52PRR3 pKa = 11.84KK4 pKa = 9.96CFEE7 pKa = 3.69GWKK10 pKa = 10.3EE11 pKa = 3.45EE12 pKa = 5.2DD13 pKa = 3.06IAKK16 pKa = 9.27MFGKK20 pKa = 9.09TDD22 pKa = 3.36TDD24 pKa = 4.91LITAEE29 pKa = 5.24DD30 pKa = 3.51IADD33 pKa = 4.46AIKK36 pKa = 10.59GKK38 pKa = 9.68KK39 pKa = 7.02QEE41 pKa = 4.46KK42 pKa = 8.43IAVYY46 pKa = 9.78KK47 pKa = 9.77QAEE50 pKa = 4.38AIKK53 pKa = 10.34KK54 pKa = 8.8GNEE57 pKa = 3.59VLTQSKK63 pKa = 10.83DD64 pKa = 3.3PASALLGMLSRR75 pKa = 11.84DD76 pKa = 3.39PNEE79 pKa = 3.82EE80 pKa = 4.07VKK82 pKa = 10.63FLSADD87 pKa = 3.31QRR89 pKa = 11.84INAIRR94 pKa = 11.84AVSKK98 pKa = 11.13AKK100 pKa = 10.46ISDD103 pKa = 3.84FMADD107 pKa = 4.01LAPTTRR113 pKa = 11.84QIFAGIATGEE123 pKa = 3.97RR124 pKa = 11.84RR125 pKa = 11.84LTKK128 pKa = 10.1SQQRR132 pKa = 11.84LLDD135 pKa = 4.09DD136 pKa = 4.96FVHH139 pKa = 6.27EE140 pKa = 4.37LYY142 pKa = 10.86GRR144 pKa = 11.84QTGNADD150 pKa = 3.31ALKK153 pKa = 10.39AAKK156 pKa = 9.05GWKK159 pKa = 9.5KK160 pKa = 9.59ATEE163 pKa = 4.15DD164 pKa = 3.23LNARR168 pKa = 11.84FGQAGGHH175 pKa = 4.99MAEE178 pKa = 4.55LDD180 pKa = 3.72DD181 pKa = 3.55WRR183 pKa = 11.84LPQKK187 pKa = 9.96HH188 pKa = 5.55NRR190 pKa = 11.84MAISKK195 pKa = 10.02AGADD199 pKa = 3.36VWVEE203 pKa = 4.13KK204 pKa = 10.91VWDD207 pKa = 4.76LIDD210 pKa = 4.04RR211 pKa = 11.84DD212 pKa = 3.91KK213 pKa = 10.96MVKK216 pKa = 10.02KK217 pKa = 10.2LRR219 pKa = 11.84KK220 pKa = 10.13GKK222 pKa = 10.38DD223 pKa = 3.07EE224 pKa = 4.88DD225 pKa = 3.79NLRR228 pKa = 11.84EE229 pKa = 4.07ALYY232 pKa = 10.62SVYY235 pKa = 11.17NNIVTDD241 pKa = 4.19GMSSSKK247 pKa = 9.54TLSKK251 pKa = 10.85KK252 pKa = 8.77FTDD255 pKa = 3.26MMRR258 pKa = 11.84SEE260 pKa = 4.39RR261 pKa = 11.84FITFKK266 pKa = 11.19DD267 pKa = 3.09SDD269 pKa = 3.55SWLKK273 pKa = 8.89YY274 pKa = 8.94QRR276 pKa = 11.84EE277 pKa = 4.12FGDD280 pKa = 3.78TNVYY284 pKa = 11.19ASMLGHH290 pKa = 7.29IDD292 pKa = 3.3NMSRR296 pKa = 11.84AIGMMEE302 pKa = 4.21TFGPDD307 pKa = 3.13PDD309 pKa = 3.64IGFNTLEE316 pKa = 4.17RR317 pKa = 11.84AVKK320 pKa = 8.58TKK322 pKa = 10.6KK323 pKa = 10.59GLTSRR328 pKa = 11.84QPTGARR334 pKa = 11.84PTFDD338 pKa = 2.99MLMGYY343 pKa = 10.95NMVEE347 pKa = 4.19EE348 pKa = 4.03QTVWGNRR355 pKa = 11.84VAGLRR360 pKa = 11.84NLWTASKK367 pKa = 10.51LGAAVVSALTDD378 pKa = 3.34SVYY381 pKa = 11.46ASMAASYY388 pKa = 11.13NAMSPARR395 pKa = 11.84VLRR398 pKa = 11.84RR399 pKa = 11.84MLSEE403 pKa = 3.8VMKK406 pKa = 9.81PSKK409 pKa = 10.78SEE411 pKa = 3.66ASRR414 pKa = 11.84KK415 pKa = 9.07LWAQDD420 pKa = 3.33FGFGAEE426 pKa = 4.04FALDD430 pKa = 3.77RR431 pKa = 11.84MAMTSDD437 pKa = 3.49YY438 pKa = 8.57TQSFGGHH445 pKa = 5.76RR446 pKa = 11.84SRR448 pKa = 11.84NLAEE452 pKa = 3.71AVMVVSGMNQWTQSARR468 pKa = 11.84ASFQFEE474 pKa = 4.0FATALTRR481 pKa = 11.84AADD484 pKa = 4.22SKK486 pKa = 11.01WSDD489 pKa = 3.69LPEE492 pKa = 4.25KK493 pKa = 9.71MRR495 pKa = 11.84NSMGRR500 pKa = 11.84YY501 pKa = 9.46GITEE505 pKa = 4.2SDD507 pKa = 3.06WAAIAAAPRR516 pKa = 11.84TNYY519 pKa = 9.85KK520 pKa = 8.69GNKK523 pKa = 8.84MIDD526 pKa = 3.95PRR528 pKa = 11.84NMDD531 pKa = 4.11AEE533 pKa = 4.43LQTKK537 pKa = 9.25LVGMVDD543 pKa = 4.38GEE545 pKa = 4.57TMMAVPTPDD554 pKa = 2.75ARR556 pKa = 11.84TRR558 pKa = 11.84AFMAGGTKK566 pKa = 10.05SGNFGGEE573 pKa = 3.73LHH575 pKa = 6.91RR576 pKa = 11.84SLFMFHH582 pKa = 7.09SFPITTIMNQWRR594 pKa = 11.84RR595 pKa = 11.84VFTGKK600 pKa = 10.49GYY602 pKa = 10.71SGAFDD607 pKa = 3.68RR608 pKa = 11.84MSAAAIMVGATSVLGVGIIQAKK630 pKa = 10.37DD631 pKa = 2.95ILNGKK636 pKa = 8.84KK637 pKa = 9.12PRR639 pKa = 11.84SMSDD643 pKa = 2.81PKK645 pKa = 11.07LWIEE649 pKa = 4.16GMAQGGSFNYY659 pKa = 10.07IGDD662 pKa = 4.36LMRR665 pKa = 11.84NAASGYY671 pKa = 7.93SHH673 pKa = 7.71DD674 pKa = 3.7MTSYY678 pKa = 10.6VGGPVLAYY686 pKa = 10.14GDD688 pKa = 4.21WVAMTAADD696 pKa = 3.96MAKK699 pKa = 10.46GDD701 pKa = 4.53AEE703 pKa = 4.16SAMARR708 pKa = 11.84TANFATQQIPFNNLWYY724 pKa = 9.66TKK726 pKa = 10.08IATDD730 pKa = 3.74RR731 pKa = 11.84LLMDD735 pKa = 5.18RR736 pKa = 11.84IRR738 pKa = 11.84RR739 pKa = 11.84LSDD742 pKa = 3.08PEE744 pKa = 3.89YY745 pKa = 11.05DD746 pKa = 3.37KK747 pKa = 11.59KK748 pKa = 11.03QLNKK752 pKa = 8.09MRR754 pKa = 11.84KK755 pKa = 5.49MQRR758 pKa = 11.84TSQQEE763 pKa = 4.44YY764 pKa = 7.04WWSPPIGGQSNIEE777 pKa = 4.28SPFEE781 pKa = 3.78EE782 pKa = 4.53
MM1 pKa = 7.52PRR3 pKa = 11.84KK4 pKa = 9.96CFEE7 pKa = 3.69GWKK10 pKa = 10.3EE11 pKa = 3.45EE12 pKa = 5.2DD13 pKa = 3.06IAKK16 pKa = 9.27MFGKK20 pKa = 9.09TDD22 pKa = 3.36TDD24 pKa = 4.91LITAEE29 pKa = 5.24DD30 pKa = 3.51IADD33 pKa = 4.46AIKK36 pKa = 10.59GKK38 pKa = 9.68KK39 pKa = 7.02QEE41 pKa = 4.46KK42 pKa = 8.43IAVYY46 pKa = 9.78KK47 pKa = 9.77QAEE50 pKa = 4.38AIKK53 pKa = 10.34KK54 pKa = 8.8GNEE57 pKa = 3.59VLTQSKK63 pKa = 10.83DD64 pKa = 3.3PASALLGMLSRR75 pKa = 11.84DD76 pKa = 3.39PNEE79 pKa = 3.82EE80 pKa = 4.07VKK82 pKa = 10.63FLSADD87 pKa = 3.31QRR89 pKa = 11.84INAIRR94 pKa = 11.84AVSKK98 pKa = 11.13AKK100 pKa = 10.46ISDD103 pKa = 3.84FMADD107 pKa = 4.01LAPTTRR113 pKa = 11.84QIFAGIATGEE123 pKa = 3.97RR124 pKa = 11.84RR125 pKa = 11.84LTKK128 pKa = 10.1SQQRR132 pKa = 11.84LLDD135 pKa = 4.09DD136 pKa = 4.96FVHH139 pKa = 6.27EE140 pKa = 4.37LYY142 pKa = 10.86GRR144 pKa = 11.84QTGNADD150 pKa = 3.31ALKK153 pKa = 10.39AAKK156 pKa = 9.05GWKK159 pKa = 9.5KK160 pKa = 9.59ATEE163 pKa = 4.15DD164 pKa = 3.23LNARR168 pKa = 11.84FGQAGGHH175 pKa = 4.99MAEE178 pKa = 4.55LDD180 pKa = 3.72DD181 pKa = 3.55WRR183 pKa = 11.84LPQKK187 pKa = 9.96HH188 pKa = 5.55NRR190 pKa = 11.84MAISKK195 pKa = 10.02AGADD199 pKa = 3.36VWVEE203 pKa = 4.13KK204 pKa = 10.91VWDD207 pKa = 4.76LIDD210 pKa = 4.04RR211 pKa = 11.84DD212 pKa = 3.91KK213 pKa = 10.96MVKK216 pKa = 10.02KK217 pKa = 10.2LRR219 pKa = 11.84KK220 pKa = 10.13GKK222 pKa = 10.38DD223 pKa = 3.07EE224 pKa = 4.88DD225 pKa = 3.79NLRR228 pKa = 11.84EE229 pKa = 4.07ALYY232 pKa = 10.62SVYY235 pKa = 11.17NNIVTDD241 pKa = 4.19GMSSSKK247 pKa = 9.54TLSKK251 pKa = 10.85KK252 pKa = 8.77FTDD255 pKa = 3.26MMRR258 pKa = 11.84SEE260 pKa = 4.39RR261 pKa = 11.84FITFKK266 pKa = 11.19DD267 pKa = 3.09SDD269 pKa = 3.55SWLKK273 pKa = 8.89YY274 pKa = 8.94QRR276 pKa = 11.84EE277 pKa = 4.12FGDD280 pKa = 3.78TNVYY284 pKa = 11.19ASMLGHH290 pKa = 7.29IDD292 pKa = 3.3NMSRR296 pKa = 11.84AIGMMEE302 pKa = 4.21TFGPDD307 pKa = 3.13PDD309 pKa = 3.64IGFNTLEE316 pKa = 4.17RR317 pKa = 11.84AVKK320 pKa = 8.58TKK322 pKa = 10.6KK323 pKa = 10.59GLTSRR328 pKa = 11.84QPTGARR334 pKa = 11.84PTFDD338 pKa = 2.99MLMGYY343 pKa = 10.95NMVEE347 pKa = 4.19EE348 pKa = 4.03QTVWGNRR355 pKa = 11.84VAGLRR360 pKa = 11.84NLWTASKK367 pKa = 10.51LGAAVVSALTDD378 pKa = 3.34SVYY381 pKa = 11.46ASMAASYY388 pKa = 11.13NAMSPARR395 pKa = 11.84VLRR398 pKa = 11.84RR399 pKa = 11.84MLSEE403 pKa = 3.8VMKK406 pKa = 9.81PSKK409 pKa = 10.78SEE411 pKa = 3.66ASRR414 pKa = 11.84KK415 pKa = 9.07LWAQDD420 pKa = 3.33FGFGAEE426 pKa = 4.04FALDD430 pKa = 3.77RR431 pKa = 11.84MAMTSDD437 pKa = 3.49YY438 pKa = 8.57TQSFGGHH445 pKa = 5.76RR446 pKa = 11.84SRR448 pKa = 11.84NLAEE452 pKa = 3.71AVMVVSGMNQWTQSARR468 pKa = 11.84ASFQFEE474 pKa = 4.0FATALTRR481 pKa = 11.84AADD484 pKa = 4.22SKK486 pKa = 11.01WSDD489 pKa = 3.69LPEE492 pKa = 4.25KK493 pKa = 9.71MRR495 pKa = 11.84NSMGRR500 pKa = 11.84YY501 pKa = 9.46GITEE505 pKa = 4.2SDD507 pKa = 3.06WAAIAAAPRR516 pKa = 11.84TNYY519 pKa = 9.85KK520 pKa = 8.69GNKK523 pKa = 8.84MIDD526 pKa = 3.95PRR528 pKa = 11.84NMDD531 pKa = 4.11AEE533 pKa = 4.43LQTKK537 pKa = 9.25LVGMVDD543 pKa = 4.38GEE545 pKa = 4.57TMMAVPTPDD554 pKa = 2.75ARR556 pKa = 11.84TRR558 pKa = 11.84AFMAGGTKK566 pKa = 10.05SGNFGGEE573 pKa = 3.73LHH575 pKa = 6.91RR576 pKa = 11.84SLFMFHH582 pKa = 7.09SFPITTIMNQWRR594 pKa = 11.84RR595 pKa = 11.84VFTGKK600 pKa = 10.49GYY602 pKa = 10.71SGAFDD607 pKa = 3.68RR608 pKa = 11.84MSAAAIMVGATSVLGVGIIQAKK630 pKa = 10.37DD631 pKa = 2.95ILNGKK636 pKa = 8.84KK637 pKa = 9.12PRR639 pKa = 11.84SMSDD643 pKa = 2.81PKK645 pKa = 11.07LWIEE649 pKa = 4.16GMAQGGSFNYY659 pKa = 10.07IGDD662 pKa = 4.36LMRR665 pKa = 11.84NAASGYY671 pKa = 7.93SHH673 pKa = 7.71DD674 pKa = 3.7MTSYY678 pKa = 10.6VGGPVLAYY686 pKa = 10.14GDD688 pKa = 4.21WVAMTAADD696 pKa = 3.96MAKK699 pKa = 10.46GDD701 pKa = 4.53AEE703 pKa = 4.16SAMARR708 pKa = 11.84TANFATQQIPFNNLWYY724 pKa = 9.66TKK726 pKa = 10.08IATDD730 pKa = 3.74RR731 pKa = 11.84LLMDD735 pKa = 5.18RR736 pKa = 11.84IRR738 pKa = 11.84RR739 pKa = 11.84LSDD742 pKa = 3.08PEE744 pKa = 3.89YY745 pKa = 11.05DD746 pKa = 3.37KK747 pKa = 11.59KK748 pKa = 11.03QLNKK752 pKa = 8.09MRR754 pKa = 11.84KK755 pKa = 5.49MQRR758 pKa = 11.84TSQQEE763 pKa = 4.44YY764 pKa = 7.04WWSPPIGGQSNIEE777 pKa = 4.28SPFEE781 pKa = 3.78EE782 pKa = 4.53
Molecular weight: 87.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5479 |
105 |
782 |
456.6 |
50.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.414 ± 0.54 | 1.04 ± 0.182 |
6.516 ± 0.385 | 6.352 ± 0.674 |
3.687 ± 0.285 | 9.034 ± 1.332 |
1.825 ± 0.29 | 5.676 ± 0.368 |
5.84 ± 0.619 | 6.881 ± 0.409 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.34 ± 0.508 | 4.636 ± 0.396 |
4.07 ± 0.259 | 3.815 ± 0.27 |
6.023 ± 0.356 | 6.059 ± 0.572 |
5.384 ± 0.431 | 5.986 ± 0.428 |
2.117 ± 0.251 | 3.304 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
