
Human papillomavirus 9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 2
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
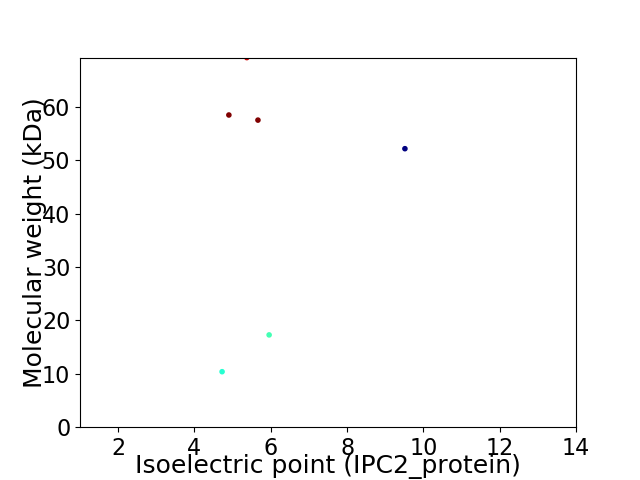
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q02480|VL1_HPV09 Major capsid protein L1 OS=Human papillomavirus 9 OX=10621 GN=L1 PE=3 SV=2
MM1 pKa = 7.4IGKK4 pKa = 8.52EE5 pKa = 3.76ATIPEE10 pKa = 4.48VVLEE14 pKa = 4.17LQEE17 pKa = 4.89LVQPTADD24 pKa = 3.45LHH26 pKa = 7.11CYY28 pKa = 10.26EE29 pKa = 5.08EE30 pKa = 4.54LTEE33 pKa = 4.52EE34 pKa = 4.17PAEE37 pKa = 4.13EE38 pKa = 4.26EE39 pKa = 4.29QCLTPYY45 pKa = 10.41KK46 pKa = 10.16IVAGCGCGARR56 pKa = 11.84LRR58 pKa = 11.84LYY60 pKa = 10.93VLATNLGIRR69 pKa = 11.84AQQEE73 pKa = 4.23LLLGDD78 pKa = 3.67IQLVCPEE85 pKa = 3.69CRR87 pKa = 11.84GRR89 pKa = 11.84LRR91 pKa = 11.84HH92 pKa = 5.58EE93 pKa = 4.03
MM1 pKa = 7.4IGKK4 pKa = 8.52EE5 pKa = 3.76ATIPEE10 pKa = 4.48VVLEE14 pKa = 4.17LQEE17 pKa = 4.89LVQPTADD24 pKa = 3.45LHH26 pKa = 7.11CYY28 pKa = 10.26EE29 pKa = 5.08EE30 pKa = 4.54LTEE33 pKa = 4.52EE34 pKa = 4.17PAEE37 pKa = 4.13EE38 pKa = 4.26EE39 pKa = 4.29QCLTPYY45 pKa = 10.41KK46 pKa = 10.16IVAGCGCGARR56 pKa = 11.84LRR58 pKa = 11.84LYY60 pKa = 10.93VLATNLGIRR69 pKa = 11.84AQQEE73 pKa = 4.23LLLGDD78 pKa = 3.67IQLVCPEE85 pKa = 3.69CRR87 pKa = 11.84GRR89 pKa = 11.84LRR91 pKa = 11.84HH92 pKa = 5.58EE93 pKa = 4.03
Molecular weight: 10.39 kDa
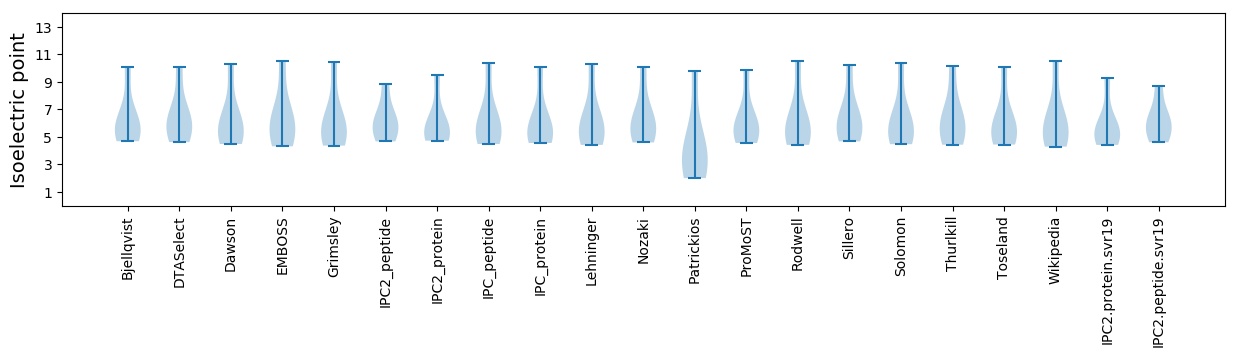
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P36801|VE6_HPV09 Protein E6 OS=Human papillomavirus 9 OX=10621 GN=E6 PE=3 SV=1
MM1 pKa = 6.95EE2 pKa = 4.43TLSARR7 pKa = 11.84FNALQEE13 pKa = 4.28TLMDD17 pKa = 4.67LYY19 pKa = 11.24EE20 pKa = 4.58SGRR23 pKa = 11.84EE24 pKa = 3.93DD25 pKa = 3.48LQSQIDD31 pKa = 3.25HH32 pKa = 6.24WQTLRR37 pKa = 11.84QEE39 pKa = 4.4QILLHH44 pKa = 5.12YY45 pKa = 9.83ARR47 pKa = 11.84KK48 pKa = 9.52NGVMRR53 pKa = 11.84LGYY56 pKa = 10.31QPVPPLATSEE66 pKa = 4.14QKK68 pKa = 11.07AKK70 pKa = 10.79DD71 pKa = 3.87AIGMVLLLQSLQRR84 pKa = 11.84SAYY87 pKa = 8.84GQEE90 pKa = 3.94PWTLAQTSLEE100 pKa = 4.19AVRR103 pKa = 11.84SPPAYY108 pKa = 9.98AFKK111 pKa = 10.67KK112 pKa = 9.46GPQNIEE118 pKa = 3.71VVYY121 pKa = 10.94DD122 pKa = 3.85GDD124 pKa = 4.11PDD126 pKa = 3.91NVMSYY131 pKa = 8.35TIWNFIYY138 pKa = 10.36YY139 pKa = 7.86QTVNDD144 pKa = 3.51TWEE147 pKa = 4.23KK148 pKa = 9.26VQGHH152 pKa = 5.4VDD154 pKa = 3.48YY155 pKa = 11.12FGAYY159 pKa = 8.98YY160 pKa = 10.73FEE162 pKa = 4.79GTVKK166 pKa = 9.88TYY168 pKa = 10.85YY169 pKa = 10.3INFDD173 pKa = 3.42KK174 pKa = 11.16DD175 pKa = 2.74AARR178 pKa = 11.84YY179 pKa = 8.73GRR181 pKa = 11.84TGVWEE186 pKa = 3.79VHH188 pKa = 4.71VNKK191 pKa = 10.43DD192 pKa = 2.96IVFAPVTSSSPPTGDD207 pKa = 2.9GGEE210 pKa = 4.37TSKK213 pKa = 9.16HH214 pKa = 4.27TLSRR218 pKa = 11.84SGSPTTSRR226 pKa = 11.84LPATTVPTGGSRR238 pKa = 11.84TSSRR242 pKa = 11.84RR243 pKa = 11.84YY244 pKa = 7.87QRR246 pKa = 11.84KK247 pKa = 9.03ASSPTTRR254 pKa = 11.84KK255 pKa = 9.36KK256 pKa = 10.32RR257 pKa = 11.84QRR259 pKa = 11.84QGEE262 pKa = 4.49GEE264 pKa = 4.26GEE266 pKa = 4.45GEE268 pKa = 4.42GEE270 pKa = 3.83EE271 pKa = 4.26TNYY274 pKa = 10.07RR275 pKa = 11.84RR276 pKa = 11.84QRR278 pKa = 11.84SRR280 pKa = 11.84SKK282 pKa = 10.89GRR284 pKa = 11.84TEE286 pKa = 3.84TEE288 pKa = 3.5RR289 pKa = 11.84GGEE292 pKa = 3.6RR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84GRR298 pKa = 11.84SSSADD303 pKa = 2.99STTPTDD309 pKa = 3.16RR310 pKa = 11.84RR311 pKa = 11.84RR312 pKa = 11.84GRR314 pKa = 11.84GGGRR318 pKa = 11.84GPTTRR323 pKa = 11.84SQSRR327 pKa = 11.84SRR329 pKa = 11.84SRR331 pKa = 11.84SHH333 pKa = 5.79SRR335 pKa = 11.84SRR337 pKa = 11.84SRR339 pKa = 11.84GGTASRR345 pKa = 11.84VGVSPDD351 pKa = 3.16EE352 pKa = 4.05VGTRR356 pKa = 11.84VRR358 pKa = 11.84SVGAGHH364 pKa = 7.27HH365 pKa = 6.02GRR367 pKa = 11.84LARR370 pKa = 11.84LLAEE374 pKa = 4.86AKK376 pKa = 10.36DD377 pKa = 3.89PPLMLLRR384 pKa = 11.84GDD386 pKa = 4.1ANVLKK391 pKa = 10.36CYY393 pKa = 10.39RR394 pKa = 11.84FRR396 pKa = 11.84EE397 pKa = 4.17RR398 pKa = 11.84KK399 pKa = 9.12KK400 pKa = 10.4KK401 pKa = 10.03RR402 pKa = 11.84GLVKK406 pKa = 10.5YY407 pKa = 10.83YY408 pKa = 9.21STTWSWVGEE417 pKa = 4.18DD418 pKa = 3.07SCDD421 pKa = 3.11RR422 pKa = 11.84VGRR425 pKa = 11.84ARR427 pKa = 11.84MILAFDD433 pKa = 4.03TYY435 pKa = 7.77EE436 pKa = 3.99HH437 pKa = 6.51RR438 pKa = 11.84QQFIRR443 pKa = 11.84TMKK446 pKa = 10.61LPPTVDD452 pKa = 2.77WSLGNVDD459 pKa = 5.38DD460 pKa = 5.05LL461 pKa = 5.06
MM1 pKa = 6.95EE2 pKa = 4.43TLSARR7 pKa = 11.84FNALQEE13 pKa = 4.28TLMDD17 pKa = 4.67LYY19 pKa = 11.24EE20 pKa = 4.58SGRR23 pKa = 11.84EE24 pKa = 3.93DD25 pKa = 3.48LQSQIDD31 pKa = 3.25HH32 pKa = 6.24WQTLRR37 pKa = 11.84QEE39 pKa = 4.4QILLHH44 pKa = 5.12YY45 pKa = 9.83ARR47 pKa = 11.84KK48 pKa = 9.52NGVMRR53 pKa = 11.84LGYY56 pKa = 10.31QPVPPLATSEE66 pKa = 4.14QKK68 pKa = 11.07AKK70 pKa = 10.79DD71 pKa = 3.87AIGMVLLLQSLQRR84 pKa = 11.84SAYY87 pKa = 8.84GQEE90 pKa = 3.94PWTLAQTSLEE100 pKa = 4.19AVRR103 pKa = 11.84SPPAYY108 pKa = 9.98AFKK111 pKa = 10.67KK112 pKa = 9.46GPQNIEE118 pKa = 3.71VVYY121 pKa = 10.94DD122 pKa = 3.85GDD124 pKa = 4.11PDD126 pKa = 3.91NVMSYY131 pKa = 8.35TIWNFIYY138 pKa = 10.36YY139 pKa = 7.86QTVNDD144 pKa = 3.51TWEE147 pKa = 4.23KK148 pKa = 9.26VQGHH152 pKa = 5.4VDD154 pKa = 3.48YY155 pKa = 11.12FGAYY159 pKa = 8.98YY160 pKa = 10.73FEE162 pKa = 4.79GTVKK166 pKa = 9.88TYY168 pKa = 10.85YY169 pKa = 10.3INFDD173 pKa = 3.42KK174 pKa = 11.16DD175 pKa = 2.74AARR178 pKa = 11.84YY179 pKa = 8.73GRR181 pKa = 11.84TGVWEE186 pKa = 3.79VHH188 pKa = 4.71VNKK191 pKa = 10.43DD192 pKa = 2.96IVFAPVTSSSPPTGDD207 pKa = 2.9GGEE210 pKa = 4.37TSKK213 pKa = 9.16HH214 pKa = 4.27TLSRR218 pKa = 11.84SGSPTTSRR226 pKa = 11.84LPATTVPTGGSRR238 pKa = 11.84TSSRR242 pKa = 11.84RR243 pKa = 11.84YY244 pKa = 7.87QRR246 pKa = 11.84KK247 pKa = 9.03ASSPTTRR254 pKa = 11.84KK255 pKa = 9.36KK256 pKa = 10.32RR257 pKa = 11.84QRR259 pKa = 11.84QGEE262 pKa = 4.49GEE264 pKa = 4.26GEE266 pKa = 4.45GEE268 pKa = 4.42GEE270 pKa = 3.83EE271 pKa = 4.26TNYY274 pKa = 10.07RR275 pKa = 11.84RR276 pKa = 11.84QRR278 pKa = 11.84SRR280 pKa = 11.84SKK282 pKa = 10.89GRR284 pKa = 11.84TEE286 pKa = 3.84TEE288 pKa = 3.5RR289 pKa = 11.84GGEE292 pKa = 3.6RR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84GRR298 pKa = 11.84SSSADD303 pKa = 2.99STTPTDD309 pKa = 3.16RR310 pKa = 11.84RR311 pKa = 11.84RR312 pKa = 11.84GRR314 pKa = 11.84GGGRR318 pKa = 11.84GPTTRR323 pKa = 11.84SQSRR327 pKa = 11.84SRR329 pKa = 11.84SRR331 pKa = 11.84SHH333 pKa = 5.79SRR335 pKa = 11.84SRR337 pKa = 11.84SRR339 pKa = 11.84GGTASRR345 pKa = 11.84VGVSPDD351 pKa = 3.16EE352 pKa = 4.05VGTRR356 pKa = 11.84VRR358 pKa = 11.84SVGAGHH364 pKa = 7.27HH365 pKa = 6.02GRR367 pKa = 11.84LARR370 pKa = 11.84LLAEE374 pKa = 4.86AKK376 pKa = 10.36DD377 pKa = 3.89PPLMLLRR384 pKa = 11.84GDD386 pKa = 4.1ANVLKK391 pKa = 10.36CYY393 pKa = 10.39RR394 pKa = 11.84FRR396 pKa = 11.84EE397 pKa = 4.17RR398 pKa = 11.84KK399 pKa = 9.12KK400 pKa = 10.4KK401 pKa = 10.03RR402 pKa = 11.84GLVKK406 pKa = 10.5YY407 pKa = 10.83YY408 pKa = 9.21STTWSWVGEE417 pKa = 4.18DD418 pKa = 3.07SCDD421 pKa = 3.11RR422 pKa = 11.84VGRR425 pKa = 11.84ARR427 pKa = 11.84MILAFDD433 pKa = 4.03TYY435 pKa = 7.77EE436 pKa = 3.99HH437 pKa = 6.51RR438 pKa = 11.84QQFIRR443 pKa = 11.84TMKK446 pKa = 10.61LPPTVDD452 pKa = 2.77WSLGNVDD459 pKa = 5.38DD460 pKa = 5.05LL461 pKa = 5.06
Molecular weight: 52.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2347 |
93 |
605 |
391.2 |
44.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.667 ± 0.366 | 2.13 ± 0.769 |
6.135 ± 0.45 | 6.562 ± 0.55 |
3.963 ± 0.663 | 6.647 ± 0.997 |
1.96 ± 0.104 | 4.602 ± 0.767 |
4.602 ± 0.813 | 8.649 ± 1.003 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.619 ± 0.292 | 3.707 ± 0.843 |
5.411 ± 0.735 | 5.198 ± 0.35 |
7.073 ± 1.461 | 7.414 ± 0.766 |
6.945 ± 0.732 | 6.732 ± 0.668 |
1.193 ± 0.316 | 3.792 ± 0.318 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
