
Blackberry virus E
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Allexivirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
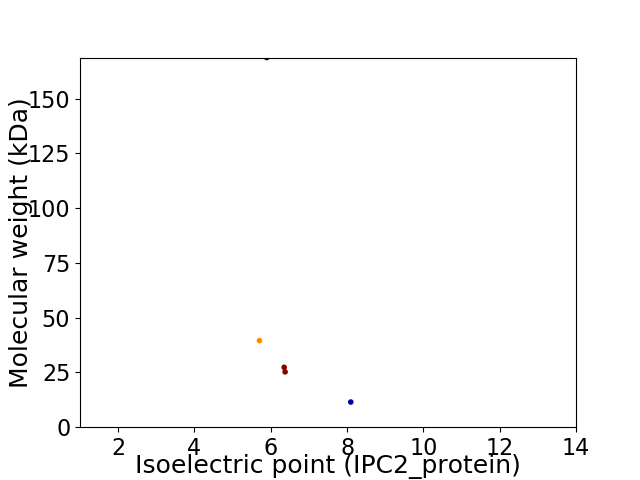
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
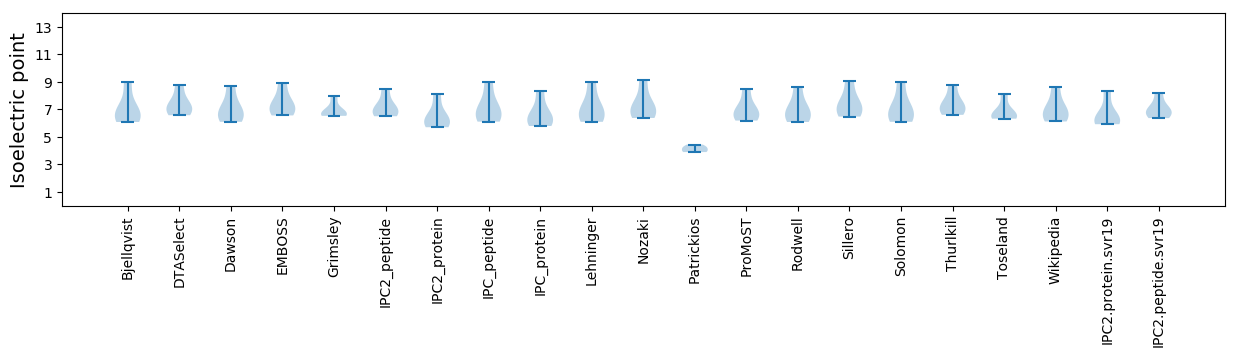
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8VAG3|F8VAG3_9VIRU Coat protein OS=Blackberry virus E OX=1043184 PE=4 SV=1
MM1 pKa = 7.67APLSVDD7 pKa = 4.78FFNQAHH13 pKa = 5.94TYY15 pKa = 9.39VVQLLEE21 pKa = 4.79RR22 pKa = 11.84IHH24 pKa = 6.71NNTIVPMGRR33 pKa = 11.84TVNRR37 pKa = 11.84IGGEE41 pKa = 3.8LDD43 pKa = 2.9QGLRR47 pKa = 11.84DD48 pKa = 3.65INAKK52 pKa = 10.26LDD54 pKa = 3.89VITSQTASAPATTVDD69 pKa = 4.29DD70 pKa = 3.97ARR72 pKa = 11.84QIPIEE77 pKa = 4.63GPASPTLTGNPLIQQTPSATRR98 pKa = 11.84FFSDD102 pKa = 3.0SNAVLSEE109 pKa = 4.04TQRR112 pKa = 11.84ITRR115 pKa = 11.84ILPSNTFPMQPSTLPTDD132 pKa = 3.53EE133 pKa = 6.19LFGQLHH139 pKa = 6.77ALHH142 pKa = 6.46QNLLQWSIDD151 pKa = 2.82IDD153 pKa = 3.66NRR155 pKa = 11.84VQQLASSVNAKK166 pKa = 10.17LDD168 pKa = 3.52AVTFNSANQVSTATLNRR185 pKa = 11.84LLEE188 pKa = 4.25EE189 pKa = 4.03LHH191 pKa = 7.51ANMATSQRR199 pKa = 11.84TGEE202 pKa = 4.13AAQTMLEE209 pKa = 4.15HH210 pKa = 6.75LADD213 pKa = 3.42IQRR216 pKa = 11.84RR217 pKa = 11.84LPSTAPSDD225 pKa = 3.92ALQQLIQLLSTNRR238 pKa = 11.84PVEE241 pKa = 4.2EE242 pKa = 4.29TTPTATPHH250 pKa = 6.27SEE252 pKa = 4.14LPALNPVHH260 pKa = 6.45NALRR264 pKa = 11.84CRR266 pKa = 11.84TYY268 pKa = 11.35GSIEE272 pKa = 4.08FNGQTFKK279 pKa = 11.24RR280 pKa = 11.84PIDD283 pKa = 3.75FVGRR287 pKa = 11.84PPSTTLHH294 pKa = 6.05LRR296 pKa = 11.84LDD298 pKa = 3.48VRR300 pKa = 11.84RR301 pKa = 11.84AADD304 pKa = 3.15HH305 pKa = 5.87TAVHH309 pKa = 6.15FTLHH313 pKa = 6.64DD314 pKa = 4.15LDD316 pKa = 4.67RR317 pKa = 11.84VLLEE321 pKa = 4.12QVIQTPHH328 pKa = 5.91QLQHH332 pKa = 6.83LPDD335 pKa = 4.94DD336 pKa = 4.24ALFLINLHH344 pKa = 6.07CPRR347 pKa = 11.84FTYY350 pKa = 10.09RR351 pKa = 11.84RR352 pKa = 11.84EE353 pKa = 4.16AGCC356 pKa = 4.32
MM1 pKa = 7.67APLSVDD7 pKa = 4.78FFNQAHH13 pKa = 5.94TYY15 pKa = 9.39VVQLLEE21 pKa = 4.79RR22 pKa = 11.84IHH24 pKa = 6.71NNTIVPMGRR33 pKa = 11.84TVNRR37 pKa = 11.84IGGEE41 pKa = 3.8LDD43 pKa = 2.9QGLRR47 pKa = 11.84DD48 pKa = 3.65INAKK52 pKa = 10.26LDD54 pKa = 3.89VITSQTASAPATTVDD69 pKa = 4.29DD70 pKa = 3.97ARR72 pKa = 11.84QIPIEE77 pKa = 4.63GPASPTLTGNPLIQQTPSATRR98 pKa = 11.84FFSDD102 pKa = 3.0SNAVLSEE109 pKa = 4.04TQRR112 pKa = 11.84ITRR115 pKa = 11.84ILPSNTFPMQPSTLPTDD132 pKa = 3.53EE133 pKa = 6.19LFGQLHH139 pKa = 6.77ALHH142 pKa = 6.46QNLLQWSIDD151 pKa = 2.82IDD153 pKa = 3.66NRR155 pKa = 11.84VQQLASSVNAKK166 pKa = 10.17LDD168 pKa = 3.52AVTFNSANQVSTATLNRR185 pKa = 11.84LLEE188 pKa = 4.25EE189 pKa = 4.03LHH191 pKa = 7.51ANMATSQRR199 pKa = 11.84TGEE202 pKa = 4.13AAQTMLEE209 pKa = 4.15HH210 pKa = 6.75LADD213 pKa = 3.42IQRR216 pKa = 11.84RR217 pKa = 11.84LPSTAPSDD225 pKa = 3.92ALQQLIQLLSTNRR238 pKa = 11.84PVEE241 pKa = 4.2EE242 pKa = 4.29TTPTATPHH250 pKa = 6.27SEE252 pKa = 4.14LPALNPVHH260 pKa = 6.45NALRR264 pKa = 11.84CRR266 pKa = 11.84TYY268 pKa = 11.35GSIEE272 pKa = 4.08FNGQTFKK279 pKa = 11.24RR280 pKa = 11.84PIDD283 pKa = 3.75FVGRR287 pKa = 11.84PPSTTLHH294 pKa = 6.05LRR296 pKa = 11.84LDD298 pKa = 3.48VRR300 pKa = 11.84RR301 pKa = 11.84AADD304 pKa = 3.15HH305 pKa = 5.87TAVHH309 pKa = 6.15FTLHH313 pKa = 6.64DD314 pKa = 4.15LDD316 pKa = 4.67RR317 pKa = 11.84VLLEE321 pKa = 4.12QVIQTPHH328 pKa = 5.91QLQHH332 pKa = 6.83LPDD335 pKa = 4.94DD336 pKa = 4.24ALFLINLHH344 pKa = 6.07CPRR347 pKa = 11.84FTYY350 pKa = 10.09RR351 pKa = 11.84RR352 pKa = 11.84EE353 pKa = 4.16AGCC356 pKa = 4.32
Molecular weight: 39.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8VAG2|F8VAG2_9VIRU Serine-rich p40 protein OS=Blackberry virus E OX=1043184 PE=4 SV=1
MM1 pKa = 7.54SFTPPPDD8 pKa = 3.42FSKK11 pKa = 11.24VYY13 pKa = 9.15LAAVAGMCATLAIHH27 pKa = 6.25IQISSRR33 pKa = 11.84RR34 pKa = 11.84PHH36 pKa = 7.14VGDD39 pKa = 4.6NIHH42 pKa = 6.52HH43 pKa = 6.77LPHH46 pKa = 6.72GGHH49 pKa = 5.67YY50 pKa = 10.0QDD52 pKa = 3.3GNKK55 pKa = 10.06RR56 pKa = 11.84IIYY59 pKa = 10.06AGSHH63 pKa = 5.81YY64 pKa = 10.93SNQLQTGDD72 pKa = 2.87HH73 pKa = 6.11WAAILVAVLILAILVSEE90 pKa = 4.41RR91 pKa = 11.84CLRR94 pKa = 11.84PSNRR98 pKa = 11.84SCPHH102 pKa = 5.11HH103 pKa = 6.53HH104 pKa = 6.96
MM1 pKa = 7.54SFTPPPDD8 pKa = 3.42FSKK11 pKa = 11.24VYY13 pKa = 9.15LAAVAGMCATLAIHH27 pKa = 6.25IQISSRR33 pKa = 11.84RR34 pKa = 11.84PHH36 pKa = 7.14VGDD39 pKa = 4.6NIHH42 pKa = 6.52HH43 pKa = 6.77LPHH46 pKa = 6.72GGHH49 pKa = 5.67YY50 pKa = 10.0QDD52 pKa = 3.3GNKK55 pKa = 10.06RR56 pKa = 11.84IIYY59 pKa = 10.06AGSHH63 pKa = 5.81YY64 pKa = 10.93SNQLQTGDD72 pKa = 2.87HH73 pKa = 6.11WAAILVAVLILAILVSEE90 pKa = 4.41RR91 pKa = 11.84CLRR94 pKa = 11.84PSNRR98 pKa = 11.84SCPHH102 pKa = 5.11HH103 pKa = 6.53HH104 pKa = 6.96
Molecular weight: 11.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2442 |
104 |
1499 |
488.4 |
54.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.681 ± 1.109 | 1.392 ± 0.349 |
5.651 ± 0.561 | 5.201 ± 0.699 |
3.931 ± 0.229 | 4.627 ± 0.361 |
3.89 ± 0.648 | 4.382 ± 0.426 |
4.545 ± 1.214 | 10.36 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.597 ± 0.241 | 4.545 ± 0.662 |
7.043 ± 0.724 | 5.119 ± 0.565 |
5.324 ± 0.481 | 6.224 ± 0.659 |
7.944 ± 0.741 | 5.856 ± 0.299 |
0.696 ± 0.116 | 2.989 ± 0.511 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
