
Hubei hepe-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.11
Get precalculated fractions of proteins
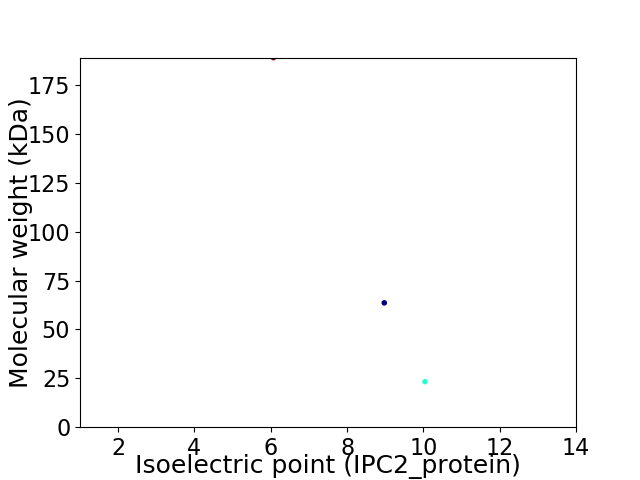
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK13|A0A1L3KK13_9VIRU Uncharacterized protein OS=Hubei hepe-like virus 1 OX=1922894 PE=4 SV=1
MM1 pKa = 7.58LGLNPASNKK10 pKa = 9.57TIIDD14 pKa = 3.61FEE16 pKa = 4.81KK17 pKa = 10.09NKK19 pKa = 10.45AFEE22 pKa = 4.06FHH24 pKa = 6.96KK25 pKa = 10.66KK26 pKa = 10.61LNGKK30 pKa = 9.72AIRR33 pKa = 11.84LPFSLKK39 pKa = 10.73AQDD42 pKa = 3.96HH43 pKa = 6.39SFLSSLYY50 pKa = 9.41APQPIEE56 pKa = 4.71LGTARR61 pKa = 11.84PPSHH65 pKa = 6.84PVVAALVRR73 pKa = 11.84IQRR76 pKa = 11.84DD77 pKa = 3.2RR78 pKa = 11.84MNSIARR84 pKa = 11.84HH85 pKa = 5.55SGHH88 pKa = 6.2NVLFFDD94 pKa = 5.16SSLSPLTDD102 pKa = 3.04NAAFIANGGNFHH114 pKa = 7.07LCLNTEE120 pKa = 4.28DD121 pKa = 4.06PRR123 pKa = 11.84YY124 pKa = 9.22HH125 pKa = 6.18QRR127 pKa = 11.84LANALFDD134 pKa = 3.71HH135 pKa = 6.85HH136 pKa = 6.44NQPGFVKK143 pKa = 10.54DD144 pKa = 3.49VVQAARR150 pKa = 11.84LGINTEE156 pKa = 4.35LVCHH160 pKa = 6.77RR161 pKa = 11.84GLHH164 pKa = 5.55HH165 pKa = 7.24CNYY168 pKa = 8.45TSPIAFAPNTISNIDD183 pKa = 3.44LTTIAKK189 pKa = 9.94LFYY192 pKa = 10.28SHH194 pKa = 7.45GIHH197 pKa = 6.23VLHH200 pKa = 7.32AFQIFPALGLVQNRR214 pKa = 11.84HH215 pKa = 5.8HH216 pKa = 6.81YY217 pKa = 10.08IDD219 pKa = 3.51YY220 pKa = 10.65QYY222 pKa = 11.09TFEE225 pKa = 5.18IDD227 pKa = 3.29NDD229 pKa = 3.37EE230 pKa = 4.23CLIGNVDD237 pKa = 4.0DD238 pKa = 4.26PSYY241 pKa = 10.85GYY243 pKa = 10.4RR244 pKa = 11.84YY245 pKa = 10.15NWTTFKK251 pKa = 10.99SHH253 pKa = 7.68FINSGIVTPFGFNILIEE270 pKa = 4.17KK271 pKa = 8.18TRR273 pKa = 11.84NHH275 pKa = 6.53GIMWEE280 pKa = 4.13LKK282 pKa = 7.19YY283 pKa = 9.59TRR285 pKa = 11.84VNGTVDD291 pKa = 3.85FPFRR295 pKa = 11.84LPIIPDD301 pKa = 3.18DD302 pKa = 3.68TMRR305 pKa = 11.84FPDD308 pKa = 3.39ICGVIQKK315 pKa = 10.09GFYY318 pKa = 10.53LSGDD322 pKa = 3.79VVSKK326 pKa = 10.9YY327 pKa = 10.78SFSGFDD333 pKa = 3.68FPPNFPFIVTSKK345 pKa = 10.38RR346 pKa = 11.84KK347 pKa = 9.9CEE349 pKa = 3.62QLYY352 pKa = 9.31QFIVTRR358 pKa = 11.84VEE360 pKa = 3.83KK361 pKa = 11.02SFDD364 pKa = 3.8LEE366 pKa = 3.92TTAAYY371 pKa = 9.96ARR373 pKa = 11.84SLLRR377 pKa = 11.84RR378 pKa = 11.84TIVGDD383 pKa = 3.7HH384 pKa = 6.56VIEE387 pKa = 4.13EE388 pKa = 4.19AWRR391 pKa = 11.84IDD393 pKa = 3.58TTEE396 pKa = 4.03LQHH399 pKa = 6.83LIVSAYY405 pKa = 8.67ILAMTSRR412 pKa = 11.84QVSNQRR418 pKa = 11.84VSAAVRR424 pKa = 11.84LLQSRR429 pKa = 11.84SSIYY433 pKa = 10.66SGVFNQIGTEE443 pKa = 3.76ILNLLGSTGARR454 pKa = 11.84LFPGIPFSFVDD465 pKa = 3.25RR466 pKa = 11.84FTRR469 pKa = 11.84YY470 pKa = 8.12TEE472 pKa = 4.8AISGRR477 pKa = 11.84SNNIFFKK484 pKa = 10.86LIEE487 pKa = 4.26NMQSFFDD494 pKa = 3.8IDD496 pKa = 3.74HH497 pKa = 6.57QIHH500 pKa = 6.05YY501 pKa = 10.02SAAFDD506 pKa = 3.61VPFIDD511 pKa = 5.86KK512 pKa = 10.86YY513 pKa = 11.06LLPYY517 pKa = 8.77PARR520 pKa = 11.84KK521 pKa = 8.63IVHH524 pKa = 6.21GLFHH528 pKa = 7.23HH529 pKa = 6.44SRR531 pKa = 11.84TLATTMMRR539 pKa = 11.84RR540 pKa = 11.84FRR542 pKa = 11.84TLADD546 pKa = 3.24TTIYY550 pKa = 10.28HH551 pKa = 5.47YY552 pKa = 10.84RR553 pKa = 11.84QFRR556 pKa = 11.84LEE558 pKa = 3.81ATLRR562 pKa = 11.84CIDD565 pKa = 5.49IEE567 pKa = 4.25EE568 pKa = 4.01QWIGRR573 pKa = 11.84LLQSYY578 pKa = 11.42ANIFPPNHH586 pKa = 6.39PKK588 pKa = 9.98VVGWRR593 pKa = 11.84LRR595 pKa = 11.84SQQWQQRR602 pKa = 11.84RR603 pKa = 11.84QVAQDD608 pKa = 3.83LIARR612 pKa = 11.84NRR614 pKa = 11.84KK615 pKa = 8.92LLISGPNKK623 pKa = 9.81PLVNLRR629 pKa = 11.84DD630 pKa = 3.64PKK632 pKa = 10.64TRR634 pKa = 11.84LIDD637 pKa = 3.75SPPVHH642 pKa = 6.36PTVITVPLLDD652 pKa = 3.5QHH654 pKa = 7.01SKK656 pKa = 10.83LLSASDD662 pKa = 4.05KK663 pKa = 10.68LAQAVDD669 pKa = 3.81DD670 pKa = 4.4RR671 pKa = 11.84LTDD674 pKa = 3.47ISNNASEE681 pKa = 5.04IFGSTCDD688 pKa = 3.69PSFLPQYY695 pKa = 7.84THH697 pKa = 6.58NCDD700 pKa = 2.77HH701 pKa = 7.19TIYY704 pKa = 10.4HH705 pKa = 5.53QVDD708 pKa = 3.77VPGDD712 pKa = 3.75GMCAFHH718 pKa = 6.62TMAHH722 pKa = 6.18FLGVNATEE730 pKa = 4.06LRR732 pKa = 11.84EE733 pKa = 3.99QIRR736 pKa = 11.84DD737 pKa = 3.32SDD739 pKa = 3.73IAPQQVVDD747 pKa = 5.03LLSDD751 pKa = 3.85PDD753 pKa = 3.22AWGDD757 pKa = 3.75YY758 pKa = 10.81PCFAVCAQIYY768 pKa = 8.51GVRR771 pKa = 11.84FCIEE775 pKa = 4.05FVDD778 pKa = 4.01SRR780 pKa = 11.84DD781 pKa = 3.51KK782 pKa = 11.23IFFLTPGNGPIYY794 pKa = 10.13HH795 pKa = 6.17VRR797 pKa = 11.84LEE799 pKa = 4.51HH800 pKa = 6.18NHH802 pKa = 5.86FSPLEE807 pKa = 3.87TDD809 pKa = 2.26IDD811 pKa = 4.25FPIEE815 pKa = 4.0GSNVPGIFNLVSSASKK831 pKa = 9.9PLPSFDD837 pKa = 4.36YY838 pKa = 11.0LDD840 pKa = 3.79PLYY843 pKa = 10.89RR844 pKa = 11.84EE845 pKa = 4.77LKK847 pKa = 9.38GQNTQLANSITARR860 pKa = 11.84LNRR863 pKa = 11.84LKK865 pKa = 9.73TAKK868 pKa = 9.4FAKK871 pKa = 10.24GEE873 pKa = 4.16IYY875 pKa = 10.5LGPPGSCKK883 pKa = 9.48TRR885 pKa = 11.84ACIKK889 pKa = 10.29KK890 pKa = 10.1LAGTGKK896 pKa = 9.99RR897 pKa = 11.84VLVSCPTRR905 pKa = 11.84ALAADD910 pKa = 4.21WEE912 pKa = 4.83NKK914 pKa = 8.84TSTEE918 pKa = 3.88DD919 pKa = 3.72FAVLTYY925 pKa = 10.07QAALVHH931 pKa = 6.44GFAEE935 pKa = 4.18QFDD938 pKa = 4.59TIVVDD943 pKa = 3.78EE944 pKa = 5.42CFTIPYY950 pKa = 10.07SYY952 pKa = 10.89ILAFAALARR961 pKa = 11.84VRR963 pKa = 11.84LILLGDD969 pKa = 3.95PNQITSIDD977 pKa = 3.73FSEE980 pKa = 4.9SKK982 pKa = 10.53CHH984 pKa = 6.3TGTPRR989 pKa = 11.84LEE991 pKa = 5.11DD992 pKa = 3.87NLDD995 pKa = 3.76CYY997 pKa = 11.41DD998 pKa = 4.75LYY1000 pKa = 11.43EE1001 pKa = 4.46CAATSRR1007 pKa = 11.84IPEE1010 pKa = 4.91DD1011 pKa = 3.26IVEE1014 pKa = 4.22TDD1016 pKa = 3.73LFQRR1020 pKa = 11.84TYY1022 pKa = 10.94PGISTTNHH1030 pKa = 5.75RR1031 pKa = 11.84KK1032 pKa = 9.37SLHH1035 pKa = 5.49IHH1037 pKa = 7.26DD1038 pKa = 4.56PTAPLPANHH1047 pKa = 5.95QVLCFTQEE1055 pKa = 3.98AKK1057 pKa = 10.84AQLCQFYY1064 pKa = 9.18PQVKK1068 pKa = 7.81TVHH1071 pKa = 6.03EE1072 pKa = 4.38AQGQTFDD1079 pKa = 4.92DD1080 pKa = 4.07VVLVVGSSPGEE1091 pKa = 3.6LGLAYY1096 pKa = 9.91EE1097 pKa = 4.92SYY1099 pKa = 10.32NHH1101 pKa = 6.71VIVACSRR1108 pKa = 11.84HH1109 pKa = 4.48TQNLVLSIDD1118 pKa = 3.87DD1119 pKa = 3.48VATFLQRR1126 pKa = 11.84TGLQPEE1132 pKa = 4.07PLVYY1136 pKa = 11.09VNDD1139 pKa = 3.85DD1140 pKa = 3.93VEE1142 pKa = 4.29LHH1144 pKa = 5.81TRR1146 pKa = 11.84DD1147 pKa = 4.88NIDD1150 pKa = 3.36DD1151 pKa = 3.85TPRR1154 pKa = 11.84IVPADD1159 pKa = 3.89YY1160 pKa = 11.35VPFSPPAVEE1169 pKa = 5.19SFADD1173 pKa = 3.77NPPSFIKK1180 pKa = 10.73DD1181 pKa = 3.26VDD1183 pKa = 3.75PSSVRR1188 pKa = 11.84EE1189 pKa = 3.83VFDD1192 pKa = 3.12IYY1194 pKa = 11.06FPVRR1198 pKa = 11.84EE1199 pKa = 4.29KK1200 pKa = 10.78QLEE1203 pKa = 3.99DD1204 pKa = 3.58CYY1206 pKa = 11.44YY1207 pKa = 10.75ATTTSYY1213 pKa = 9.18NTEE1216 pKa = 3.81GSGRR1220 pKa = 11.84ATVQFEE1226 pKa = 4.27KK1227 pKa = 10.79VAEE1230 pKa = 4.09NSLQEE1235 pKa = 3.92MRR1237 pKa = 11.84TFTRR1241 pKa = 11.84STFSIDD1247 pKa = 2.91PRR1249 pKa = 11.84GRR1251 pKa = 11.84YY1252 pKa = 4.06TTSRR1256 pKa = 11.84SRR1258 pKa = 11.84IQEE1261 pKa = 3.62VHH1263 pKa = 5.69TALQRR1268 pKa = 11.84LLKK1271 pKa = 9.78FTMNPPRR1278 pKa = 11.84AYY1280 pKa = 10.44VLEE1283 pKa = 4.07HH1284 pKa = 6.91SIRR1287 pKa = 11.84LVNQLNDD1294 pKa = 5.36LIPFCPVQTRR1304 pKa = 11.84NPEE1307 pKa = 4.71LIHH1310 pKa = 5.41QAYY1313 pKa = 9.7VEE1315 pKa = 4.39YY1316 pKa = 10.28ASKK1319 pKa = 8.18MTEE1322 pKa = 3.63RR1323 pKa = 11.84GQGIEE1328 pKa = 5.15DD1329 pKa = 3.38ITEE1332 pKa = 3.86KK1333 pKa = 10.54TIIDD1337 pKa = 4.38PGTVSLFLKK1346 pKa = 10.33QQFKK1350 pKa = 10.68QDD1352 pKa = 3.68YY1353 pKa = 8.47TSGEE1357 pKa = 3.73GALRR1361 pKa = 11.84DD1362 pKa = 3.89KK1363 pKa = 11.02SGQPISAWDD1372 pKa = 3.32KK1373 pKa = 7.76RR1374 pKa = 11.84TNFIFSVWARR1384 pKa = 11.84ILEE1387 pKa = 4.38FYY1389 pKa = 11.01LLDD1392 pKa = 3.98SPTPRR1397 pKa = 11.84FKK1399 pKa = 10.78FFSRR1403 pKa = 11.84VSEE1406 pKa = 4.06PVLLKK1411 pKa = 11.02CLDD1414 pKa = 3.91SFPKK1418 pKa = 10.59DD1419 pKa = 3.47GISYY1423 pKa = 8.94FCNDD1427 pKa = 2.64FTEE1430 pKa = 4.7YY1431 pKa = 10.89DD1432 pKa = 3.4SCQNEE1437 pKa = 4.31LEE1439 pKa = 4.26HH1440 pKa = 7.14EE1441 pKa = 4.18ILAEE1445 pKa = 3.81VLRR1448 pKa = 11.84RR1449 pKa = 11.84IGAPAALIEE1458 pKa = 4.78HH1459 pKa = 6.48YY1460 pKa = 10.89LHH1462 pKa = 6.74IRR1464 pKa = 11.84RR1465 pKa = 11.84NRR1467 pKa = 11.84VAYY1470 pKa = 10.05SQTVKK1475 pKa = 10.85CKK1477 pKa = 10.53VKK1479 pKa = 10.43NKK1481 pKa = 9.49RR1482 pKa = 11.84DD1483 pKa = 3.44SGEE1486 pKa = 3.98PFTLIGNTIFTSCIVLDD1503 pKa = 4.02IMDD1506 pKa = 5.33DD1507 pKa = 3.76FDD1509 pKa = 4.55FLLVKK1514 pKa = 10.68GDD1516 pKa = 3.95DD1517 pKa = 4.1SAAGGYY1523 pKa = 9.48NISVRR1528 pKa = 11.84HH1529 pKa = 5.64EE1530 pKa = 4.44KK1531 pKa = 9.5IAHH1534 pKa = 4.99YY1535 pKa = 9.41FQTNRR1540 pKa = 11.84YY1541 pKa = 7.21VMKK1544 pKa = 10.29PNVSDD1549 pKa = 3.22SCEE1552 pKa = 3.67FVGYY1556 pKa = 8.09ITNVNGTCLDD1566 pKa = 3.23IVRR1569 pKa = 11.84LCARR1573 pKa = 11.84LVGRR1577 pKa = 11.84KK1578 pKa = 9.06FKK1580 pKa = 11.07DD1581 pKa = 3.2EE1582 pKa = 4.29EE1583 pKa = 4.29DD1584 pKa = 3.99FNDD1587 pKa = 3.55YY1588 pKa = 10.61VISVRR1593 pKa = 11.84DD1594 pKa = 3.48RR1595 pKa = 11.84VRR1597 pKa = 11.84SFASVDD1603 pKa = 3.21LMHH1606 pKa = 7.24RR1607 pKa = 11.84SIAVISLHH1615 pKa = 6.04YY1616 pKa = 10.79KK1617 pKa = 8.17MAPEE1621 pKa = 3.93HH1622 pKa = 6.79VEE1624 pKa = 3.38WLYY1627 pKa = 11.67NYY1629 pKa = 10.39LLNFAYY1635 pKa = 10.7NKK1637 pKa = 10.12IPFDD1641 pKa = 3.82ALMSSKK1647 pKa = 10.59QSIWNLISNIPTRR1660 pKa = 11.84FF1661 pKa = 3.17
MM1 pKa = 7.58LGLNPASNKK10 pKa = 9.57TIIDD14 pKa = 3.61FEE16 pKa = 4.81KK17 pKa = 10.09NKK19 pKa = 10.45AFEE22 pKa = 4.06FHH24 pKa = 6.96KK25 pKa = 10.66KK26 pKa = 10.61LNGKK30 pKa = 9.72AIRR33 pKa = 11.84LPFSLKK39 pKa = 10.73AQDD42 pKa = 3.96HH43 pKa = 6.39SFLSSLYY50 pKa = 9.41APQPIEE56 pKa = 4.71LGTARR61 pKa = 11.84PPSHH65 pKa = 6.84PVVAALVRR73 pKa = 11.84IQRR76 pKa = 11.84DD77 pKa = 3.2RR78 pKa = 11.84MNSIARR84 pKa = 11.84HH85 pKa = 5.55SGHH88 pKa = 6.2NVLFFDD94 pKa = 5.16SSLSPLTDD102 pKa = 3.04NAAFIANGGNFHH114 pKa = 7.07LCLNTEE120 pKa = 4.28DD121 pKa = 4.06PRR123 pKa = 11.84YY124 pKa = 9.22HH125 pKa = 6.18QRR127 pKa = 11.84LANALFDD134 pKa = 3.71HH135 pKa = 6.85HH136 pKa = 6.44NQPGFVKK143 pKa = 10.54DD144 pKa = 3.49VVQAARR150 pKa = 11.84LGINTEE156 pKa = 4.35LVCHH160 pKa = 6.77RR161 pKa = 11.84GLHH164 pKa = 5.55HH165 pKa = 7.24CNYY168 pKa = 8.45TSPIAFAPNTISNIDD183 pKa = 3.44LTTIAKK189 pKa = 9.94LFYY192 pKa = 10.28SHH194 pKa = 7.45GIHH197 pKa = 6.23VLHH200 pKa = 7.32AFQIFPALGLVQNRR214 pKa = 11.84HH215 pKa = 5.8HH216 pKa = 6.81YY217 pKa = 10.08IDD219 pKa = 3.51YY220 pKa = 10.65QYY222 pKa = 11.09TFEE225 pKa = 5.18IDD227 pKa = 3.29NDD229 pKa = 3.37EE230 pKa = 4.23CLIGNVDD237 pKa = 4.0DD238 pKa = 4.26PSYY241 pKa = 10.85GYY243 pKa = 10.4RR244 pKa = 11.84YY245 pKa = 10.15NWTTFKK251 pKa = 10.99SHH253 pKa = 7.68FINSGIVTPFGFNILIEE270 pKa = 4.17KK271 pKa = 8.18TRR273 pKa = 11.84NHH275 pKa = 6.53GIMWEE280 pKa = 4.13LKK282 pKa = 7.19YY283 pKa = 9.59TRR285 pKa = 11.84VNGTVDD291 pKa = 3.85FPFRR295 pKa = 11.84LPIIPDD301 pKa = 3.18DD302 pKa = 3.68TMRR305 pKa = 11.84FPDD308 pKa = 3.39ICGVIQKK315 pKa = 10.09GFYY318 pKa = 10.53LSGDD322 pKa = 3.79VVSKK326 pKa = 10.9YY327 pKa = 10.78SFSGFDD333 pKa = 3.68FPPNFPFIVTSKK345 pKa = 10.38RR346 pKa = 11.84KK347 pKa = 9.9CEE349 pKa = 3.62QLYY352 pKa = 9.31QFIVTRR358 pKa = 11.84VEE360 pKa = 3.83KK361 pKa = 11.02SFDD364 pKa = 3.8LEE366 pKa = 3.92TTAAYY371 pKa = 9.96ARR373 pKa = 11.84SLLRR377 pKa = 11.84RR378 pKa = 11.84TIVGDD383 pKa = 3.7HH384 pKa = 6.56VIEE387 pKa = 4.13EE388 pKa = 4.19AWRR391 pKa = 11.84IDD393 pKa = 3.58TTEE396 pKa = 4.03LQHH399 pKa = 6.83LIVSAYY405 pKa = 8.67ILAMTSRR412 pKa = 11.84QVSNQRR418 pKa = 11.84VSAAVRR424 pKa = 11.84LLQSRR429 pKa = 11.84SSIYY433 pKa = 10.66SGVFNQIGTEE443 pKa = 3.76ILNLLGSTGARR454 pKa = 11.84LFPGIPFSFVDD465 pKa = 3.25RR466 pKa = 11.84FTRR469 pKa = 11.84YY470 pKa = 8.12TEE472 pKa = 4.8AISGRR477 pKa = 11.84SNNIFFKK484 pKa = 10.86LIEE487 pKa = 4.26NMQSFFDD494 pKa = 3.8IDD496 pKa = 3.74HH497 pKa = 6.57QIHH500 pKa = 6.05YY501 pKa = 10.02SAAFDD506 pKa = 3.61VPFIDD511 pKa = 5.86KK512 pKa = 10.86YY513 pKa = 11.06LLPYY517 pKa = 8.77PARR520 pKa = 11.84KK521 pKa = 8.63IVHH524 pKa = 6.21GLFHH528 pKa = 7.23HH529 pKa = 6.44SRR531 pKa = 11.84TLATTMMRR539 pKa = 11.84RR540 pKa = 11.84FRR542 pKa = 11.84TLADD546 pKa = 3.24TTIYY550 pKa = 10.28HH551 pKa = 5.47YY552 pKa = 10.84RR553 pKa = 11.84QFRR556 pKa = 11.84LEE558 pKa = 3.81ATLRR562 pKa = 11.84CIDD565 pKa = 5.49IEE567 pKa = 4.25EE568 pKa = 4.01QWIGRR573 pKa = 11.84LLQSYY578 pKa = 11.42ANIFPPNHH586 pKa = 6.39PKK588 pKa = 9.98VVGWRR593 pKa = 11.84LRR595 pKa = 11.84SQQWQQRR602 pKa = 11.84RR603 pKa = 11.84QVAQDD608 pKa = 3.83LIARR612 pKa = 11.84NRR614 pKa = 11.84KK615 pKa = 8.92LLISGPNKK623 pKa = 9.81PLVNLRR629 pKa = 11.84DD630 pKa = 3.64PKK632 pKa = 10.64TRR634 pKa = 11.84LIDD637 pKa = 3.75SPPVHH642 pKa = 6.36PTVITVPLLDD652 pKa = 3.5QHH654 pKa = 7.01SKK656 pKa = 10.83LLSASDD662 pKa = 4.05KK663 pKa = 10.68LAQAVDD669 pKa = 3.81DD670 pKa = 4.4RR671 pKa = 11.84LTDD674 pKa = 3.47ISNNASEE681 pKa = 5.04IFGSTCDD688 pKa = 3.69PSFLPQYY695 pKa = 7.84THH697 pKa = 6.58NCDD700 pKa = 2.77HH701 pKa = 7.19TIYY704 pKa = 10.4HH705 pKa = 5.53QVDD708 pKa = 3.77VPGDD712 pKa = 3.75GMCAFHH718 pKa = 6.62TMAHH722 pKa = 6.18FLGVNATEE730 pKa = 4.06LRR732 pKa = 11.84EE733 pKa = 3.99QIRR736 pKa = 11.84DD737 pKa = 3.32SDD739 pKa = 3.73IAPQQVVDD747 pKa = 5.03LLSDD751 pKa = 3.85PDD753 pKa = 3.22AWGDD757 pKa = 3.75YY758 pKa = 10.81PCFAVCAQIYY768 pKa = 8.51GVRR771 pKa = 11.84FCIEE775 pKa = 4.05FVDD778 pKa = 4.01SRR780 pKa = 11.84DD781 pKa = 3.51KK782 pKa = 11.23IFFLTPGNGPIYY794 pKa = 10.13HH795 pKa = 6.17VRR797 pKa = 11.84LEE799 pKa = 4.51HH800 pKa = 6.18NHH802 pKa = 5.86FSPLEE807 pKa = 3.87TDD809 pKa = 2.26IDD811 pKa = 4.25FPIEE815 pKa = 4.0GSNVPGIFNLVSSASKK831 pKa = 9.9PLPSFDD837 pKa = 4.36YY838 pKa = 11.0LDD840 pKa = 3.79PLYY843 pKa = 10.89RR844 pKa = 11.84EE845 pKa = 4.77LKK847 pKa = 9.38GQNTQLANSITARR860 pKa = 11.84LNRR863 pKa = 11.84LKK865 pKa = 9.73TAKK868 pKa = 9.4FAKK871 pKa = 10.24GEE873 pKa = 4.16IYY875 pKa = 10.5LGPPGSCKK883 pKa = 9.48TRR885 pKa = 11.84ACIKK889 pKa = 10.29KK890 pKa = 10.1LAGTGKK896 pKa = 9.99RR897 pKa = 11.84VLVSCPTRR905 pKa = 11.84ALAADD910 pKa = 4.21WEE912 pKa = 4.83NKK914 pKa = 8.84TSTEE918 pKa = 3.88DD919 pKa = 3.72FAVLTYY925 pKa = 10.07QAALVHH931 pKa = 6.44GFAEE935 pKa = 4.18QFDD938 pKa = 4.59TIVVDD943 pKa = 3.78EE944 pKa = 5.42CFTIPYY950 pKa = 10.07SYY952 pKa = 10.89ILAFAALARR961 pKa = 11.84VRR963 pKa = 11.84LILLGDD969 pKa = 3.95PNQITSIDD977 pKa = 3.73FSEE980 pKa = 4.9SKK982 pKa = 10.53CHH984 pKa = 6.3TGTPRR989 pKa = 11.84LEE991 pKa = 5.11DD992 pKa = 3.87NLDD995 pKa = 3.76CYY997 pKa = 11.41DD998 pKa = 4.75LYY1000 pKa = 11.43EE1001 pKa = 4.46CAATSRR1007 pKa = 11.84IPEE1010 pKa = 4.91DD1011 pKa = 3.26IVEE1014 pKa = 4.22TDD1016 pKa = 3.73LFQRR1020 pKa = 11.84TYY1022 pKa = 10.94PGISTTNHH1030 pKa = 5.75RR1031 pKa = 11.84KK1032 pKa = 9.37SLHH1035 pKa = 5.49IHH1037 pKa = 7.26DD1038 pKa = 4.56PTAPLPANHH1047 pKa = 5.95QVLCFTQEE1055 pKa = 3.98AKK1057 pKa = 10.84AQLCQFYY1064 pKa = 9.18PQVKK1068 pKa = 7.81TVHH1071 pKa = 6.03EE1072 pKa = 4.38AQGQTFDD1079 pKa = 4.92DD1080 pKa = 4.07VVLVVGSSPGEE1091 pKa = 3.6LGLAYY1096 pKa = 9.91EE1097 pKa = 4.92SYY1099 pKa = 10.32NHH1101 pKa = 6.71VIVACSRR1108 pKa = 11.84HH1109 pKa = 4.48TQNLVLSIDD1118 pKa = 3.87DD1119 pKa = 3.48VATFLQRR1126 pKa = 11.84TGLQPEE1132 pKa = 4.07PLVYY1136 pKa = 11.09VNDD1139 pKa = 3.85DD1140 pKa = 3.93VEE1142 pKa = 4.29LHH1144 pKa = 5.81TRR1146 pKa = 11.84DD1147 pKa = 4.88NIDD1150 pKa = 3.36DD1151 pKa = 3.85TPRR1154 pKa = 11.84IVPADD1159 pKa = 3.89YY1160 pKa = 11.35VPFSPPAVEE1169 pKa = 5.19SFADD1173 pKa = 3.77NPPSFIKK1180 pKa = 10.73DD1181 pKa = 3.26VDD1183 pKa = 3.75PSSVRR1188 pKa = 11.84EE1189 pKa = 3.83VFDD1192 pKa = 3.12IYY1194 pKa = 11.06FPVRR1198 pKa = 11.84EE1199 pKa = 4.29KK1200 pKa = 10.78QLEE1203 pKa = 3.99DD1204 pKa = 3.58CYY1206 pKa = 11.44YY1207 pKa = 10.75ATTTSYY1213 pKa = 9.18NTEE1216 pKa = 3.81GSGRR1220 pKa = 11.84ATVQFEE1226 pKa = 4.27KK1227 pKa = 10.79VAEE1230 pKa = 4.09NSLQEE1235 pKa = 3.92MRR1237 pKa = 11.84TFTRR1241 pKa = 11.84STFSIDD1247 pKa = 2.91PRR1249 pKa = 11.84GRR1251 pKa = 11.84YY1252 pKa = 4.06TTSRR1256 pKa = 11.84SRR1258 pKa = 11.84IQEE1261 pKa = 3.62VHH1263 pKa = 5.69TALQRR1268 pKa = 11.84LLKK1271 pKa = 9.78FTMNPPRR1278 pKa = 11.84AYY1280 pKa = 10.44VLEE1283 pKa = 4.07HH1284 pKa = 6.91SIRR1287 pKa = 11.84LVNQLNDD1294 pKa = 5.36LIPFCPVQTRR1304 pKa = 11.84NPEE1307 pKa = 4.71LIHH1310 pKa = 5.41QAYY1313 pKa = 9.7VEE1315 pKa = 4.39YY1316 pKa = 10.28ASKK1319 pKa = 8.18MTEE1322 pKa = 3.63RR1323 pKa = 11.84GQGIEE1328 pKa = 5.15DD1329 pKa = 3.38ITEE1332 pKa = 3.86KK1333 pKa = 10.54TIIDD1337 pKa = 4.38PGTVSLFLKK1346 pKa = 10.33QQFKK1350 pKa = 10.68QDD1352 pKa = 3.68YY1353 pKa = 8.47TSGEE1357 pKa = 3.73GALRR1361 pKa = 11.84DD1362 pKa = 3.89KK1363 pKa = 11.02SGQPISAWDD1372 pKa = 3.32KK1373 pKa = 7.76RR1374 pKa = 11.84TNFIFSVWARR1384 pKa = 11.84ILEE1387 pKa = 4.38FYY1389 pKa = 11.01LLDD1392 pKa = 3.98SPTPRR1397 pKa = 11.84FKK1399 pKa = 10.78FFSRR1403 pKa = 11.84VSEE1406 pKa = 4.06PVLLKK1411 pKa = 11.02CLDD1414 pKa = 3.91SFPKK1418 pKa = 10.59DD1419 pKa = 3.47GISYY1423 pKa = 8.94FCNDD1427 pKa = 2.64FTEE1430 pKa = 4.7YY1431 pKa = 10.89DD1432 pKa = 3.4SCQNEE1437 pKa = 4.31LEE1439 pKa = 4.26HH1440 pKa = 7.14EE1441 pKa = 4.18ILAEE1445 pKa = 3.81VLRR1448 pKa = 11.84RR1449 pKa = 11.84IGAPAALIEE1458 pKa = 4.78HH1459 pKa = 6.48YY1460 pKa = 10.89LHH1462 pKa = 6.74IRR1464 pKa = 11.84RR1465 pKa = 11.84NRR1467 pKa = 11.84VAYY1470 pKa = 10.05SQTVKK1475 pKa = 10.85CKK1477 pKa = 10.53VKK1479 pKa = 10.43NKK1481 pKa = 9.49RR1482 pKa = 11.84DD1483 pKa = 3.44SGEE1486 pKa = 3.98PFTLIGNTIFTSCIVLDD1503 pKa = 4.02IMDD1506 pKa = 5.33DD1507 pKa = 3.76FDD1509 pKa = 4.55FLLVKK1514 pKa = 10.68GDD1516 pKa = 3.95DD1517 pKa = 4.1SAAGGYY1523 pKa = 9.48NISVRR1528 pKa = 11.84HH1529 pKa = 5.64EE1530 pKa = 4.44KK1531 pKa = 9.5IAHH1534 pKa = 4.99YY1535 pKa = 9.41FQTNRR1540 pKa = 11.84YY1541 pKa = 7.21VMKK1544 pKa = 10.29PNVSDD1549 pKa = 3.22SCEE1552 pKa = 3.67FVGYY1556 pKa = 8.09ITNVNGTCLDD1566 pKa = 3.23IVRR1569 pKa = 11.84LCARR1573 pKa = 11.84LVGRR1577 pKa = 11.84KK1578 pKa = 9.06FKK1580 pKa = 11.07DD1581 pKa = 3.2EE1582 pKa = 4.29EE1583 pKa = 4.29DD1584 pKa = 3.99FNDD1587 pKa = 3.55YY1588 pKa = 10.61VISVRR1593 pKa = 11.84DD1594 pKa = 3.48RR1595 pKa = 11.84VRR1597 pKa = 11.84SFASVDD1603 pKa = 3.21LMHH1606 pKa = 7.24RR1607 pKa = 11.84SIAVISLHH1615 pKa = 6.04YY1616 pKa = 10.79KK1617 pKa = 8.17MAPEE1621 pKa = 3.93HH1622 pKa = 6.79VEE1624 pKa = 3.38WLYY1627 pKa = 11.67NYY1629 pKa = 10.39LLNFAYY1635 pKa = 10.7NKK1637 pKa = 10.12IPFDD1641 pKa = 3.82ALMSSKK1647 pKa = 10.59QSIWNLISNIPTRR1660 pKa = 11.84FF1661 pKa = 3.17
Molecular weight: 189.08 kDa
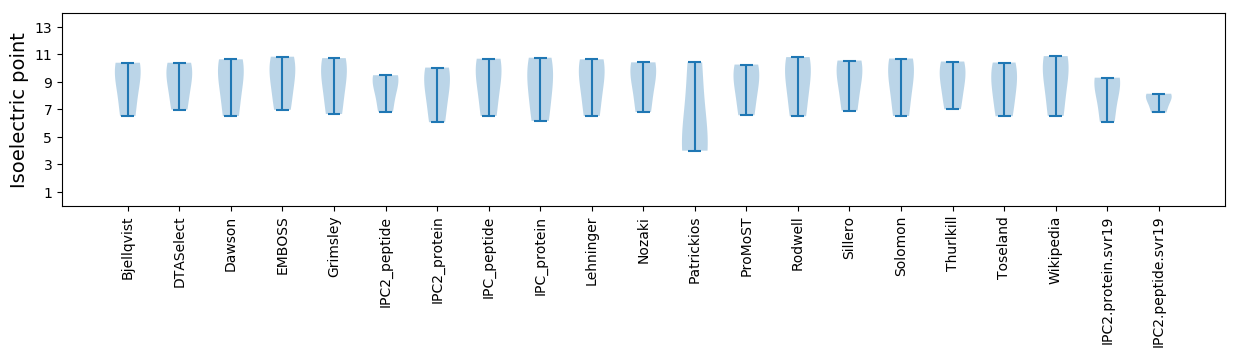
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KK49|A0A1L3KK49_9VIRU DiSB-ORF2_chro domain-containing protein OS=Hubei hepe-like virus 1 OX=1922894 PE=4 SV=1
MM1 pKa = 7.24SFSRR5 pKa = 11.84ASFRR9 pKa = 11.84GRR11 pKa = 11.84FRR13 pKa = 11.84GRR15 pKa = 11.84GRR17 pKa = 11.84AAFMRR22 pKa = 11.84PPPSYY27 pKa = 10.12FYY29 pKa = 10.87YY30 pKa = 10.21PPPQPLSVPIRR41 pKa = 11.84PAPPPPTNGNAPKK54 pKa = 10.05QGYY57 pKa = 9.85LEE59 pKa = 4.84TILASCQEE67 pKa = 4.4NISNPFRR74 pKa = 11.84LALFILGACFVVAWAINASSPGHH97 pKa = 5.33QVVQHH102 pKa = 5.12LRR104 pKa = 11.84GRR106 pKa = 11.84LSEE109 pKa = 4.02THH111 pKa = 4.9ATKK114 pKa = 10.82VKK116 pKa = 10.0TITDD120 pKa = 4.59FLTTNHH126 pKa = 5.19VRR128 pKa = 11.84FFASGVMLIPFSLLYY143 pKa = 10.47ARR145 pKa = 11.84QKK147 pKa = 9.07WLTWALAFLLVIWVAPTYY165 pKa = 10.39SEE167 pKa = 3.69WQYY170 pKa = 11.75VVISLLLFAYY180 pKa = 8.93YY181 pKa = 10.12AQKK184 pKa = 9.79TKK186 pKa = 10.32EE187 pKa = 3.92NRR189 pKa = 11.84QTALVILLVGFVAIHH204 pKa = 5.96FVV206 pKa = 3.14
MM1 pKa = 7.24SFSRR5 pKa = 11.84ASFRR9 pKa = 11.84GRR11 pKa = 11.84FRR13 pKa = 11.84GRR15 pKa = 11.84GRR17 pKa = 11.84AAFMRR22 pKa = 11.84PPPSYY27 pKa = 10.12FYY29 pKa = 10.87YY30 pKa = 10.21PPPQPLSVPIRR41 pKa = 11.84PAPPPPTNGNAPKK54 pKa = 10.05QGYY57 pKa = 9.85LEE59 pKa = 4.84TILASCQEE67 pKa = 4.4NISNPFRR74 pKa = 11.84LALFILGACFVVAWAINASSPGHH97 pKa = 5.33QVVQHH102 pKa = 5.12LRR104 pKa = 11.84GRR106 pKa = 11.84LSEE109 pKa = 4.02THH111 pKa = 4.9ATKK114 pKa = 10.82VKK116 pKa = 10.0TITDD120 pKa = 4.59FLTTNHH126 pKa = 5.19VRR128 pKa = 11.84FFASGVMLIPFSLLYY143 pKa = 10.47ARR145 pKa = 11.84QKK147 pKa = 9.07WLTWALAFLLVIWVAPTYY165 pKa = 10.39SEE167 pKa = 3.69WQYY170 pKa = 11.75VVISLLLFAYY180 pKa = 8.93YY181 pKa = 10.12AQKK184 pKa = 9.79TKK186 pKa = 10.32EE187 pKa = 3.92NRR189 pKa = 11.84QTALVILLVGFVAIHH204 pKa = 5.96FVV206 pKa = 3.14
Molecular weight: 23.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2427 |
206 |
1661 |
809.0 |
92.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.18 ± 0.81 | 1.978 ± 0.169 |
5.027 ± 1.76 | 3.832 ± 0.887 |
6.757 ± 0.591 | 4.079 ± 0.683 |
3.708 ± 0.341 | 6.51 ± 0.563 |
3.214 ± 0.942 | 10.919 ± 2.624 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.071 ± 0.073 | 4.326 ± 0.584 |
6.634 ± 1.232 | 3.749 ± 0.555 |
6.345 ± 0.108 | 8.241 ± 1.685 |
6.634 ± 0.533 | 6.098 ± 0.663 |
1.03 ± 0.381 | 3.667 ± 0.442 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
