
Georgenia soli
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Bogoriellaceae; Georgenia
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
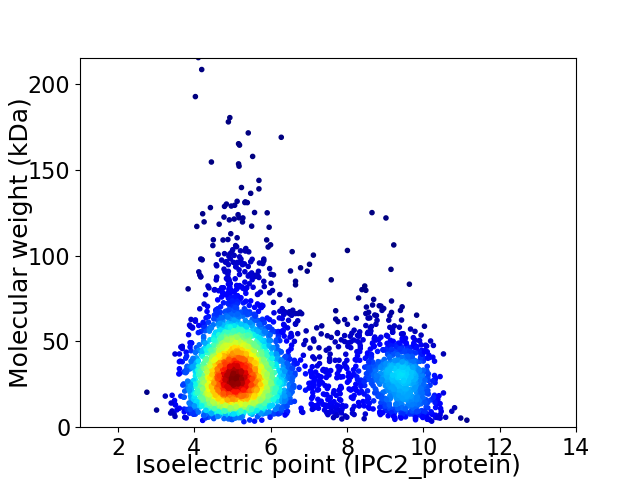
Virtual 2D-PAGE plot for 3768 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
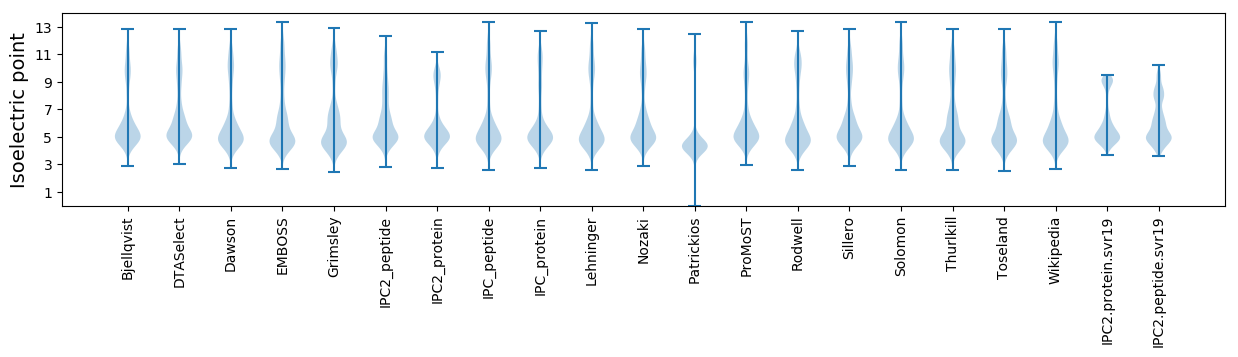
* You can choose from 21 different methods for calculating isoelectric point
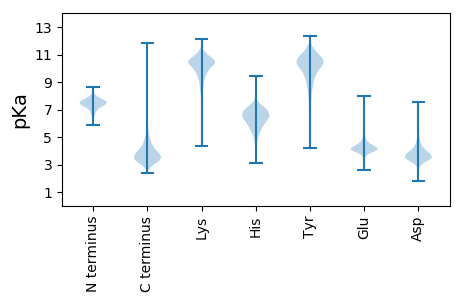
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9ELZ2|A0A2A9ELZ2_9MICO Uracil-xanthine permease OS=Georgenia soli OX=638953 GN=ATJ97_1783 PE=3 SV=1
MM1 pKa = 7.33HH2 pKa = 7.82RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84LLASTAAAAAAVLALSACAPSDD27 pKa = 3.82DD28 pKa = 4.96APDD31 pKa = 4.47PGADD35 pKa = 3.48GGATSAAGSDD45 pKa = 3.51EE46 pKa = 4.11CAEE49 pKa = 4.08PVALDD54 pKa = 3.21VWSWRR59 pKa = 11.84TEE61 pKa = 4.01DD62 pKa = 3.37KK63 pKa = 10.33DD64 pKa = 3.65TYY66 pKa = 9.8EE67 pKa = 4.17TIFDD71 pKa = 3.89VFEE74 pKa = 4.18EE75 pKa = 4.55ANPCITVNFQAYY87 pKa = 9.78KK88 pKa = 8.16NTEE91 pKa = 3.98YY92 pKa = 11.18NQILQTGLTGSDD104 pKa = 3.85GPDD107 pKa = 3.0VAQVRR112 pKa = 11.84SYY114 pKa = 11.78GLLQPLVEE122 pKa = 4.97GGNLAPLDD130 pKa = 4.07DD131 pKa = 4.61VVPALADD138 pKa = 3.64FDD140 pKa = 4.41EE141 pKa = 4.86AALDD145 pKa = 3.82GARR148 pKa = 11.84GKK150 pKa = 10.59EE151 pKa = 3.73DD152 pKa = 2.93GGIYY156 pKa = 10.09GVPFAAQTIQVFYY169 pKa = 9.23NTAIFEE175 pKa = 4.29EE176 pKa = 4.92HH177 pKa = 6.45GLEE180 pKa = 4.96EE181 pKa = 4.69PEE183 pKa = 3.92TWEE186 pKa = 5.36DD187 pKa = 4.81FIAANDD193 pKa = 3.92TLKK196 pKa = 10.87EE197 pKa = 4.09AGVTPLAVGAKK208 pKa = 9.59DD209 pKa = 2.93AWFVPTVHH217 pKa = 7.4DD218 pKa = 4.61ALTAAQYY225 pKa = 11.1GGAEE229 pKa = 4.22FQQKK233 pKa = 10.02LRR235 pKa = 11.84DD236 pKa = 3.71GEE238 pKa = 4.36TDD240 pKa = 3.81FNDD243 pKa = 3.47PAYY246 pKa = 10.55VKK248 pKa = 10.29SIQSFADD255 pKa = 3.65LEE257 pKa = 4.39QYY259 pKa = 10.07MPDD262 pKa = 3.58DD263 pKa = 4.14VVGVAYY269 pKa = 9.65TDD271 pKa = 3.51AQVLFTSGAAAMFAGGSYY289 pKa = 10.39EE290 pKa = 4.06LAFFQSTNPEE300 pKa = 3.94LEE302 pKa = 4.13LGVFQVPPPEE312 pKa = 5.08GSALDD317 pKa = 4.07HH318 pKa = 6.52PVSPGYY324 pKa = 11.09ADD326 pKa = 4.44GNWGLSAASDD336 pKa = 3.71NPEE339 pKa = 3.82EE340 pKa = 5.83AEE342 pKa = 4.48TLLNWLASEE351 pKa = 4.69EE352 pKa = 4.04FGQMVANDD360 pKa = 4.74LKK362 pKa = 10.97QFSPVPGVTFDD373 pKa = 3.65EE374 pKa = 4.79PLMQEE379 pKa = 3.64MWDD382 pKa = 4.21LYY384 pKa = 10.58QEE386 pKa = 4.29NPAPYY391 pKa = 10.13LLLVDD396 pKa = 3.99YY397 pKa = 10.64RR398 pKa = 11.84YY399 pKa = 10.78GDD401 pKa = 3.57PVGTDD406 pKa = 3.06LMGEE410 pKa = 4.77GGQQLFLGEE419 pKa = 4.14LDD421 pKa = 3.7AAGVASKK428 pKa = 10.53IQDD431 pKa = 3.74GLSAWWTPGQQ441 pKa = 3.68
MM1 pKa = 7.33HH2 pKa = 7.82RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84LLASTAAAAAAVLALSACAPSDD27 pKa = 3.82DD28 pKa = 4.96APDD31 pKa = 4.47PGADD35 pKa = 3.48GGATSAAGSDD45 pKa = 3.51EE46 pKa = 4.11CAEE49 pKa = 4.08PVALDD54 pKa = 3.21VWSWRR59 pKa = 11.84TEE61 pKa = 4.01DD62 pKa = 3.37KK63 pKa = 10.33DD64 pKa = 3.65TYY66 pKa = 9.8EE67 pKa = 4.17TIFDD71 pKa = 3.89VFEE74 pKa = 4.18EE75 pKa = 4.55ANPCITVNFQAYY87 pKa = 9.78KK88 pKa = 8.16NTEE91 pKa = 3.98YY92 pKa = 11.18NQILQTGLTGSDD104 pKa = 3.85GPDD107 pKa = 3.0VAQVRR112 pKa = 11.84SYY114 pKa = 11.78GLLQPLVEE122 pKa = 4.97GGNLAPLDD130 pKa = 4.07DD131 pKa = 4.61VVPALADD138 pKa = 3.64FDD140 pKa = 4.41EE141 pKa = 4.86AALDD145 pKa = 3.82GARR148 pKa = 11.84GKK150 pKa = 10.59EE151 pKa = 3.73DD152 pKa = 2.93GGIYY156 pKa = 10.09GVPFAAQTIQVFYY169 pKa = 9.23NTAIFEE175 pKa = 4.29EE176 pKa = 4.92HH177 pKa = 6.45GLEE180 pKa = 4.96EE181 pKa = 4.69PEE183 pKa = 3.92TWEE186 pKa = 5.36DD187 pKa = 4.81FIAANDD193 pKa = 3.92TLKK196 pKa = 10.87EE197 pKa = 4.09AGVTPLAVGAKK208 pKa = 9.59DD209 pKa = 2.93AWFVPTVHH217 pKa = 7.4DD218 pKa = 4.61ALTAAQYY225 pKa = 11.1GGAEE229 pKa = 4.22FQQKK233 pKa = 10.02LRR235 pKa = 11.84DD236 pKa = 3.71GEE238 pKa = 4.36TDD240 pKa = 3.81FNDD243 pKa = 3.47PAYY246 pKa = 10.55VKK248 pKa = 10.29SIQSFADD255 pKa = 3.65LEE257 pKa = 4.39QYY259 pKa = 10.07MPDD262 pKa = 3.58DD263 pKa = 4.14VVGVAYY269 pKa = 9.65TDD271 pKa = 3.51AQVLFTSGAAAMFAGGSYY289 pKa = 10.39EE290 pKa = 4.06LAFFQSTNPEE300 pKa = 3.94LEE302 pKa = 4.13LGVFQVPPPEE312 pKa = 5.08GSALDD317 pKa = 4.07HH318 pKa = 6.52PVSPGYY324 pKa = 11.09ADD326 pKa = 4.44GNWGLSAASDD336 pKa = 3.71NPEE339 pKa = 3.82EE340 pKa = 5.83AEE342 pKa = 4.48TLLNWLASEE351 pKa = 4.69EE352 pKa = 4.04FGQMVANDD360 pKa = 4.74LKK362 pKa = 10.97QFSPVPGVTFDD373 pKa = 3.65EE374 pKa = 4.79PLMQEE379 pKa = 3.64MWDD382 pKa = 4.21LYY384 pKa = 10.58QEE386 pKa = 4.29NPAPYY391 pKa = 10.13LLLVDD396 pKa = 3.99YY397 pKa = 10.64RR398 pKa = 11.84YY399 pKa = 10.78GDD401 pKa = 3.57PVGTDD406 pKa = 3.06LMGEE410 pKa = 4.77GGQQLFLGEE419 pKa = 4.14LDD421 pKa = 3.7AAGVASKK428 pKa = 10.53IQDD431 pKa = 3.74GLSAWWTPGQQ441 pKa = 3.68
Molecular weight: 47.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9EQ30|A0A2A9EQ30_9MICO Uncharacterized protein (DUF1697 family) OS=Georgenia soli OX=638953 GN=ATJ97_3620 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1259096 |
29 |
2069 |
334.2 |
35.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.005 ± 0.058 | 0.541 ± 0.009 |
6.103 ± 0.037 | 5.941 ± 0.042 |
2.547 ± 0.023 | 9.516 ± 0.037 |
2.133 ± 0.02 | 3.079 ± 0.031 |
1.569 ± 0.026 | 10.265 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.789 ± 0.016 | 1.664 ± 0.02 |
5.934 ± 0.034 | 2.665 ± 0.021 |
7.918 ± 0.047 | 4.952 ± 0.028 |
6.22 ± 0.031 | 9.829 ± 0.04 |
1.457 ± 0.017 | 1.874 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
