
Pectobacterium phage DU_PP_III
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Astrithrvirus; unclassified Astrithrvirus
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
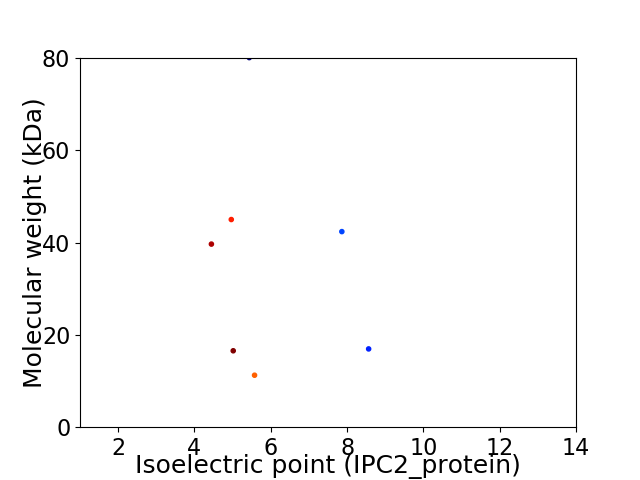
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2W5Y8|A0A2D2W5Y8_9CAUD DNA encapsidation protein OS=Pectobacterium phage DU_PP_III OX=2041490 GN=P9AB12kb_p002 PE=4 SV=1
MM1 pKa = 7.65AYY3 pKa = 10.09DD4 pKa = 3.82YY5 pKa = 10.62KK6 pKa = 11.13AKK8 pKa = 10.23RR9 pKa = 11.84ANILTLNKK17 pKa = 10.21YY18 pKa = 8.66MLAKK22 pKa = 8.04TVSMFEE28 pKa = 4.12YY29 pKa = 10.04EE30 pKa = 4.65GLPEE34 pKa = 4.63TIPYY38 pKa = 10.13EE39 pKa = 4.0EE40 pKa = 4.91LEE42 pKa = 5.65KK43 pKa = 10.33IIQQNGYY50 pKa = 10.96AFITEE55 pKa = 4.4HH56 pKa = 7.02DD57 pKa = 3.81GKK59 pKa = 10.91LYY61 pKa = 11.08AFAGSLGGEE70 pKa = 3.97LDD72 pKa = 3.95VYY74 pKa = 11.26GNPQDD79 pKa = 3.42ITITNTALNLSKK91 pKa = 9.62TFKK94 pKa = 10.55VATDD98 pKa = 3.71GVLIRR103 pKa = 11.84NDD105 pKa = 3.31DD106 pKa = 3.55MRR108 pKa = 11.84IGLMPLYY115 pKa = 10.27EE116 pKa = 4.31KK117 pKa = 11.21ANTFLVEE124 pKa = 3.78NDD126 pKa = 3.42INMMMWGYY134 pKa = 10.9NSRR137 pKa = 11.84SQKK140 pKa = 10.52VISAPDD146 pKa = 3.55DD147 pKa = 3.64KK148 pKa = 11.19TKK150 pKa = 11.18EE151 pKa = 3.98NADD154 pKa = 3.96LYY156 pKa = 10.87LKK158 pKa = 10.71KK159 pKa = 10.79LIDD162 pKa = 3.88GDD164 pKa = 3.54ISVIGEE170 pKa = 3.81NALFDD175 pKa = 4.76GIKK178 pKa = 10.82VNGSSNSQTATVTQMVEE195 pKa = 3.54LQQYY199 pKa = 9.77IKK201 pKa = 10.87ASLFNEE207 pKa = 4.19VGISANFNMKK217 pKa = 10.09RR218 pKa = 11.84EE219 pKa = 3.97RR220 pKa = 11.84LVSGEE225 pKa = 3.91VDD227 pKa = 3.12QSEE230 pKa = 4.73DD231 pKa = 3.44SLFPFVYY238 pKa = 11.02NMMKK242 pKa = 10.29CRR244 pKa = 11.84IKK246 pKa = 10.94GIEE249 pKa = 4.07ALNAKK254 pKa = 10.23YY255 pKa = 10.38EE256 pKa = 4.2LTVKK260 pKa = 10.32VDD262 pKa = 5.28FGSIWHH268 pKa = 6.76IKK270 pKa = 9.77NKK272 pKa = 10.2EE273 pKa = 4.05LVDD276 pKa = 5.48DD277 pKa = 5.34IIDD280 pKa = 4.16DD281 pKa = 4.05KK282 pKa = 11.41EE283 pKa = 4.51LPDD286 pKa = 4.06EE287 pKa = 4.64LKK289 pKa = 10.48QDD291 pKa = 3.74GTSDD295 pKa = 4.75GNNQPDD301 pKa = 3.92NNGGTGNEE309 pKa = 4.35GEE311 pKa = 4.5GEE313 pKa = 4.13RR314 pKa = 11.84EE315 pKa = 3.81RR316 pKa = 11.84GQGQATDD323 pKa = 3.63AEE325 pKa = 4.99TVAEE329 pKa = 4.3LQAIIADD336 pKa = 3.72EE337 pKa = 4.14SLPEE341 pKa = 4.09SEE343 pKa = 5.26RR344 pKa = 11.84EE345 pKa = 3.85AAKK348 pKa = 10.54EE349 pKa = 3.49LLAEE353 pKa = 4.57LGEE356 pKa = 4.33
MM1 pKa = 7.65AYY3 pKa = 10.09DD4 pKa = 3.82YY5 pKa = 10.62KK6 pKa = 11.13AKK8 pKa = 10.23RR9 pKa = 11.84ANILTLNKK17 pKa = 10.21YY18 pKa = 8.66MLAKK22 pKa = 8.04TVSMFEE28 pKa = 4.12YY29 pKa = 10.04EE30 pKa = 4.65GLPEE34 pKa = 4.63TIPYY38 pKa = 10.13EE39 pKa = 4.0EE40 pKa = 4.91LEE42 pKa = 5.65KK43 pKa = 10.33IIQQNGYY50 pKa = 10.96AFITEE55 pKa = 4.4HH56 pKa = 7.02DD57 pKa = 3.81GKK59 pKa = 10.91LYY61 pKa = 11.08AFAGSLGGEE70 pKa = 3.97LDD72 pKa = 3.95VYY74 pKa = 11.26GNPQDD79 pKa = 3.42ITITNTALNLSKK91 pKa = 9.62TFKK94 pKa = 10.55VATDD98 pKa = 3.71GVLIRR103 pKa = 11.84NDD105 pKa = 3.31DD106 pKa = 3.55MRR108 pKa = 11.84IGLMPLYY115 pKa = 10.27EE116 pKa = 4.31KK117 pKa = 11.21ANTFLVEE124 pKa = 3.78NDD126 pKa = 3.42INMMMWGYY134 pKa = 10.9NSRR137 pKa = 11.84SQKK140 pKa = 10.52VISAPDD146 pKa = 3.55DD147 pKa = 3.64KK148 pKa = 11.19TKK150 pKa = 11.18EE151 pKa = 3.98NADD154 pKa = 3.96LYY156 pKa = 10.87LKK158 pKa = 10.71KK159 pKa = 10.79LIDD162 pKa = 3.88GDD164 pKa = 3.54ISVIGEE170 pKa = 3.81NALFDD175 pKa = 4.76GIKK178 pKa = 10.82VNGSSNSQTATVTQMVEE195 pKa = 3.54LQQYY199 pKa = 9.77IKK201 pKa = 10.87ASLFNEE207 pKa = 4.19VGISANFNMKK217 pKa = 10.09RR218 pKa = 11.84EE219 pKa = 3.97RR220 pKa = 11.84LVSGEE225 pKa = 3.91VDD227 pKa = 3.12QSEE230 pKa = 4.73DD231 pKa = 3.44SLFPFVYY238 pKa = 11.02NMMKK242 pKa = 10.29CRR244 pKa = 11.84IKK246 pKa = 10.94GIEE249 pKa = 4.07ALNAKK254 pKa = 10.23YY255 pKa = 10.38EE256 pKa = 4.2LTVKK260 pKa = 10.32VDD262 pKa = 5.28FGSIWHH268 pKa = 6.76IKK270 pKa = 9.77NKK272 pKa = 10.2EE273 pKa = 4.05LVDD276 pKa = 5.48DD277 pKa = 5.34IIDD280 pKa = 4.16DD281 pKa = 4.05KK282 pKa = 11.41EE283 pKa = 4.51LPDD286 pKa = 4.06EE287 pKa = 4.64LKK289 pKa = 10.48QDD291 pKa = 3.74GTSDD295 pKa = 4.75GNNQPDD301 pKa = 3.92NNGGTGNEE309 pKa = 4.35GEE311 pKa = 4.5GEE313 pKa = 4.13RR314 pKa = 11.84EE315 pKa = 3.81RR316 pKa = 11.84GQGQATDD323 pKa = 3.63AEE325 pKa = 4.99TVAEE329 pKa = 4.3LQAIIADD336 pKa = 3.72EE337 pKa = 4.14SLPEE341 pKa = 4.09SEE343 pKa = 5.26RR344 pKa = 11.84EE345 pKa = 3.85AAKK348 pKa = 10.54EE349 pKa = 3.49LLAEE353 pKa = 4.57LGEE356 pKa = 4.33
Molecular weight: 39.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2W5Y1|A0A2D2W5Y1_9CAUD Uncharacterized protein OS=Pectobacterium phage DU_PP_III OX=2041490 GN=P9AB12kb_p007 PE=4 SV=1
MM1 pKa = 7.5IKK3 pKa = 10.41LAEE6 pKa = 4.31TYY8 pKa = 11.19LNINVDD14 pKa = 3.75GKK16 pKa = 10.89LKK18 pKa = 10.92LMEE21 pKa = 4.42YY22 pKa = 10.61YY23 pKa = 11.11NNNCYY28 pKa = 10.24QFIKK32 pKa = 10.46KK33 pKa = 8.88SRR35 pKa = 11.84KK36 pKa = 9.31YY37 pKa = 10.25KK38 pKa = 10.46IKK40 pKa = 11.12ANDD43 pKa = 3.02NWCAMFTSVIAHH55 pKa = 6.64KK56 pKa = 10.03MGLSPDD62 pKa = 3.55VFPYY66 pKa = 9.89EE67 pKa = 4.12VSVGEE72 pKa = 4.09QVKK75 pKa = 9.91LARR78 pKa = 11.84EE79 pKa = 4.08RR80 pKa = 11.84GSFTQNVEE88 pKa = 3.96LAKK91 pKa = 10.95SGDD94 pKa = 3.76LIIFNWNGDD103 pKa = 3.25AWPDD107 pKa = 3.18HH108 pKa = 6.11VGFVKK113 pKa = 10.53SVNNGIITTVEE124 pKa = 3.43GNYY127 pKa = 10.32RR128 pKa = 11.84KK129 pKa = 7.78TVGNRR134 pKa = 11.84HH135 pKa = 5.76IAMNSPFIVGIIQLL149 pKa = 3.82
MM1 pKa = 7.5IKK3 pKa = 10.41LAEE6 pKa = 4.31TYY8 pKa = 11.19LNINVDD14 pKa = 3.75GKK16 pKa = 10.89LKK18 pKa = 10.92LMEE21 pKa = 4.42YY22 pKa = 10.61YY23 pKa = 11.11NNNCYY28 pKa = 10.24QFIKK32 pKa = 10.46KK33 pKa = 8.88SRR35 pKa = 11.84KK36 pKa = 9.31YY37 pKa = 10.25KK38 pKa = 10.46IKK40 pKa = 11.12ANDD43 pKa = 3.02NWCAMFTSVIAHH55 pKa = 6.64KK56 pKa = 10.03MGLSPDD62 pKa = 3.55VFPYY66 pKa = 9.89EE67 pKa = 4.12VSVGEE72 pKa = 4.09QVKK75 pKa = 9.91LARR78 pKa = 11.84EE79 pKa = 4.08RR80 pKa = 11.84GSFTQNVEE88 pKa = 3.96LAKK91 pKa = 10.95SGDD94 pKa = 3.76LIIFNWNGDD103 pKa = 3.25AWPDD107 pKa = 3.18HH108 pKa = 6.11VGFVKK113 pKa = 10.53SVNNGIITTVEE124 pKa = 3.43GNYY127 pKa = 10.32RR128 pKa = 11.84KK129 pKa = 7.78TVGNRR134 pKa = 11.84HH135 pKa = 5.76IAMNSPFIVGIIQLL149 pKa = 3.82
Molecular weight: 16.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2213 |
103 |
690 |
316.1 |
35.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.92 ± 0.913 | 0.949 ± 0.29 |
6.507 ± 0.298 | 7.049 ± 0.567 |
4.745 ± 0.392 | 6.01 ± 0.401 |
1.446 ± 0.247 | 7.23 ± 0.394 |
8.36 ± 0.58 | 7.998 ± 0.58 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.802 ± 0.273 | 6.869 ± 0.483 |
2.35 ± 0.231 | 2.576 ± 0.371 |
3.028 ± 0.235 | 6.236 ± 0.53 |
6.236 ± 0.326 | 7.094 ± 0.669 |
1.085 ± 0.181 | 5.513 ± 0.82 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
