
Pricia antarctica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Pricia
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
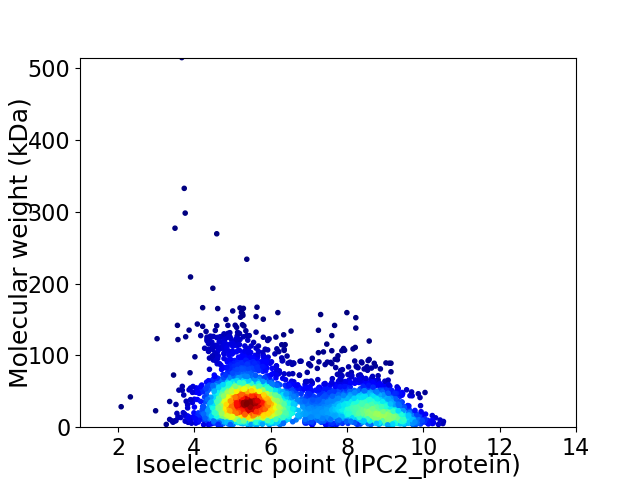
Virtual 2D-PAGE plot for 4172 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6ZS75|A0A1G6ZS75_9FLAO Uncharacterized protein OS=Pricia antarctica OX=641691 GN=SAMN05421636_10377 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.17IGVLIIGIFFMGNYY16 pKa = 10.34ALFGQITVDD25 pKa = 3.28DD26 pKa = 3.71SGYY29 pKa = 9.06TVEE32 pKa = 4.23QLVRR36 pKa = 11.84DD37 pKa = 3.68VLINSNCAATSSYY50 pKa = 11.58SSFTGTEE57 pKa = 3.8QNVNGIGYY65 pKa = 9.38FNANATDD72 pKa = 4.07FPYY75 pKa = 10.73KK76 pKa = 10.35EE77 pKa = 4.73GIILSTGRR85 pKa = 11.84ARR87 pKa = 11.84DD88 pKa = 3.56AQGPNTGINDD98 pKa = 3.89SGTEE102 pKa = 4.01SWPGDD107 pKa = 3.27QDD109 pKa = 5.54LITVTNTGNLFNATYY124 pKa = 9.92IQFDD128 pKa = 4.21FVPLTNSINFNFLFASEE145 pKa = 5.16EE146 pKa = 4.03YY147 pKa = 10.3QEE149 pKa = 5.66NYY151 pKa = 8.85QCTYY155 pKa = 9.97SDD157 pKa = 3.39VFAFILTDD165 pKa = 3.51SQGVSTNLAIVPGTEE180 pKa = 3.81EE181 pKa = 4.11PVRR184 pKa = 11.84ATTIRR189 pKa = 11.84PGIQGQCDD197 pKa = 3.28ARR199 pKa = 11.84NIDD202 pKa = 4.83FFGKK206 pKa = 10.52LNGANDD212 pKa = 4.88PISFYY217 pKa = 11.36GQTEE221 pKa = 4.26SLKK224 pKa = 10.92AEE226 pKa = 4.18SLVTPGEE233 pKa = 4.32SYY235 pKa = 10.28TIKK238 pKa = 10.78LVISDD243 pKa = 4.12NQDD246 pKa = 3.18SQVDD250 pKa = 3.72SAVFLEE256 pKa = 4.74AGSFSLGYY264 pKa = 10.5DD265 pKa = 3.1LGEE268 pKa = 4.72DD269 pKa = 2.89RR270 pKa = 11.84TVANGNPACIGEE282 pKa = 4.72NIPLDD287 pKa = 3.56ATVEE291 pKa = 4.33GVQDD295 pKa = 3.56YY296 pKa = 10.56RR297 pKa = 11.84WYY299 pKa = 10.86KK300 pKa = 10.72DD301 pKa = 3.29GSEE304 pKa = 4.18VAQWAGNPQVTLTEE318 pKa = 4.15NGQYY322 pKa = 10.1RR323 pKa = 11.84VDD325 pKa = 5.02LIFSASCISDD335 pKa = 3.1GGLTVEE341 pKa = 5.8FIVPPKK347 pKa = 10.57INGNPVDD354 pKa = 3.88ITACDD359 pKa = 3.85LDD361 pKa = 5.15GDD363 pKa = 4.17GSEE366 pKa = 4.56TFDD369 pKa = 4.02FSTNGTRR376 pKa = 11.84MMGTQDD382 pKa = 3.16SAVYY386 pKa = 9.83GVTYY390 pKa = 9.88FASEE394 pKa = 3.42VDD396 pKa = 3.48AQNFTNPIEE405 pKa = 4.34NPEE408 pKa = 3.98NYY410 pKa = 8.96EE411 pKa = 3.99VQGTEE416 pKa = 3.8TIYY419 pKa = 11.26ARR421 pKa = 11.84LSSGQSCSEE430 pKa = 3.64IVPFQLKK437 pKa = 9.66VQGLDD442 pKa = 3.55FEE444 pKa = 5.16SSLKK448 pKa = 10.6ASYY451 pKa = 10.16ILCFDD456 pKa = 3.87GEE458 pKa = 4.67GEE460 pKa = 4.15KK461 pKa = 10.95LEE463 pKa = 4.49PLPVLDD469 pKa = 4.58TGLSLSKK476 pKa = 11.42YY477 pKa = 7.58MFTWYY482 pKa = 9.59TEE484 pKa = 4.41TISEE488 pKa = 4.27EE489 pKa = 3.91NRR491 pKa = 11.84ITDD494 pKa = 3.2ATGPSFTASALGTYY508 pKa = 9.63HH509 pKa = 7.21VLLQNLEE516 pKa = 4.43FGCEE520 pKa = 3.67FSIATEE526 pKa = 4.09VLPSQQPEE534 pKa = 4.08VFEE537 pKa = 4.35VDD539 pKa = 4.1FVSDD543 pKa = 4.7LFSDD547 pKa = 3.93SNTVEE552 pKa = 3.99IKK554 pKa = 10.68AQGEE558 pKa = 4.43STYY561 pKa = 11.15LFAVDD566 pKa = 5.47DD567 pKa = 4.34SDD569 pKa = 5.03FGTSNRR575 pKa = 11.84FEE577 pKa = 4.57DD578 pKa = 4.49LSAGEE583 pKa = 4.05HH584 pKa = 4.66VAYY587 pKa = 9.56VTDD590 pKa = 4.14SEE592 pKa = 4.57NCSVLSEE599 pKa = 4.57DD600 pKa = 4.14FLVVDD605 pKa = 5.02FPRR608 pKa = 11.84FFTPNGDD615 pKa = 3.64GNNDD619 pKa = 2.2IWQIVGFPEE628 pKa = 3.88IEE630 pKa = 4.02KK631 pKa = 11.05AEE633 pKa = 3.95IAIFDD638 pKa = 3.48QYY640 pKa = 10.64GTLLYY645 pKa = 10.53QFNNGSGWDD654 pKa = 3.7GTANDD659 pKa = 4.12HH660 pKa = 6.93RR661 pKa = 11.84MPASDD666 pKa = 3.3YY667 pKa = 8.7WFRR670 pKa = 11.84IAYY673 pKa = 9.94VKK675 pKa = 11.12DD676 pKa = 3.5NVQKK680 pKa = 10.34EE681 pKa = 4.62FKK683 pKa = 10.39SHH685 pKa = 6.98FSLKK689 pKa = 10.32RR690 pKa = 3.28
MM1 pKa = 7.76KK2 pKa = 10.17IGVLIIGIFFMGNYY16 pKa = 10.34ALFGQITVDD25 pKa = 3.28DD26 pKa = 3.71SGYY29 pKa = 9.06TVEE32 pKa = 4.23QLVRR36 pKa = 11.84DD37 pKa = 3.68VLINSNCAATSSYY50 pKa = 11.58SSFTGTEE57 pKa = 3.8QNVNGIGYY65 pKa = 9.38FNANATDD72 pKa = 4.07FPYY75 pKa = 10.73KK76 pKa = 10.35EE77 pKa = 4.73GIILSTGRR85 pKa = 11.84ARR87 pKa = 11.84DD88 pKa = 3.56AQGPNTGINDD98 pKa = 3.89SGTEE102 pKa = 4.01SWPGDD107 pKa = 3.27QDD109 pKa = 5.54LITVTNTGNLFNATYY124 pKa = 9.92IQFDD128 pKa = 4.21FVPLTNSINFNFLFASEE145 pKa = 5.16EE146 pKa = 4.03YY147 pKa = 10.3QEE149 pKa = 5.66NYY151 pKa = 8.85QCTYY155 pKa = 9.97SDD157 pKa = 3.39VFAFILTDD165 pKa = 3.51SQGVSTNLAIVPGTEE180 pKa = 3.81EE181 pKa = 4.11PVRR184 pKa = 11.84ATTIRR189 pKa = 11.84PGIQGQCDD197 pKa = 3.28ARR199 pKa = 11.84NIDD202 pKa = 4.83FFGKK206 pKa = 10.52LNGANDD212 pKa = 4.88PISFYY217 pKa = 11.36GQTEE221 pKa = 4.26SLKK224 pKa = 10.92AEE226 pKa = 4.18SLVTPGEE233 pKa = 4.32SYY235 pKa = 10.28TIKK238 pKa = 10.78LVISDD243 pKa = 4.12NQDD246 pKa = 3.18SQVDD250 pKa = 3.72SAVFLEE256 pKa = 4.74AGSFSLGYY264 pKa = 10.5DD265 pKa = 3.1LGEE268 pKa = 4.72DD269 pKa = 2.89RR270 pKa = 11.84TVANGNPACIGEE282 pKa = 4.72NIPLDD287 pKa = 3.56ATVEE291 pKa = 4.33GVQDD295 pKa = 3.56YY296 pKa = 10.56RR297 pKa = 11.84WYY299 pKa = 10.86KK300 pKa = 10.72DD301 pKa = 3.29GSEE304 pKa = 4.18VAQWAGNPQVTLTEE318 pKa = 4.15NGQYY322 pKa = 10.1RR323 pKa = 11.84VDD325 pKa = 5.02LIFSASCISDD335 pKa = 3.1GGLTVEE341 pKa = 5.8FIVPPKK347 pKa = 10.57INGNPVDD354 pKa = 3.88ITACDD359 pKa = 3.85LDD361 pKa = 5.15GDD363 pKa = 4.17GSEE366 pKa = 4.56TFDD369 pKa = 4.02FSTNGTRR376 pKa = 11.84MMGTQDD382 pKa = 3.16SAVYY386 pKa = 9.83GVTYY390 pKa = 9.88FASEE394 pKa = 3.42VDD396 pKa = 3.48AQNFTNPIEE405 pKa = 4.34NPEE408 pKa = 3.98NYY410 pKa = 8.96EE411 pKa = 3.99VQGTEE416 pKa = 3.8TIYY419 pKa = 11.26ARR421 pKa = 11.84LSSGQSCSEE430 pKa = 3.64IVPFQLKK437 pKa = 9.66VQGLDD442 pKa = 3.55FEE444 pKa = 5.16SSLKK448 pKa = 10.6ASYY451 pKa = 10.16ILCFDD456 pKa = 3.87GEE458 pKa = 4.67GEE460 pKa = 4.15KK461 pKa = 10.95LEE463 pKa = 4.49PLPVLDD469 pKa = 4.58TGLSLSKK476 pKa = 11.42YY477 pKa = 7.58MFTWYY482 pKa = 9.59TEE484 pKa = 4.41TISEE488 pKa = 4.27EE489 pKa = 3.91NRR491 pKa = 11.84ITDD494 pKa = 3.2ATGPSFTASALGTYY508 pKa = 9.63HH509 pKa = 7.21VLLQNLEE516 pKa = 4.43FGCEE520 pKa = 3.67FSIATEE526 pKa = 4.09VLPSQQPEE534 pKa = 4.08VFEE537 pKa = 4.35VDD539 pKa = 4.1FVSDD543 pKa = 4.7LFSDD547 pKa = 3.93SNTVEE552 pKa = 3.99IKK554 pKa = 10.68AQGEE558 pKa = 4.43STYY561 pKa = 11.15LFAVDD566 pKa = 5.47DD567 pKa = 4.34SDD569 pKa = 5.03FGTSNRR575 pKa = 11.84FEE577 pKa = 4.57DD578 pKa = 4.49LSAGEE583 pKa = 4.05HH584 pKa = 4.66VAYY587 pKa = 9.56VTDD590 pKa = 4.14SEE592 pKa = 4.57NCSVLSEE599 pKa = 4.57DD600 pKa = 4.14FLVVDD605 pKa = 5.02FPRR608 pKa = 11.84FFTPNGDD615 pKa = 3.64GNNDD619 pKa = 2.2IWQIVGFPEE628 pKa = 3.88IEE630 pKa = 4.02KK631 pKa = 11.05AEE633 pKa = 3.95IAIFDD638 pKa = 3.48QYY640 pKa = 10.64GTLLYY645 pKa = 10.53QFNNGSGWDD654 pKa = 3.7GTANDD659 pKa = 4.12HH660 pKa = 6.93RR661 pKa = 11.84MPASDD666 pKa = 3.3YY667 pKa = 8.7WFRR670 pKa = 11.84IAYY673 pKa = 9.94VKK675 pKa = 11.12DD676 pKa = 3.5NVQKK680 pKa = 10.34EE681 pKa = 4.62FKK683 pKa = 10.39SHH685 pKa = 6.98FSLKK689 pKa = 10.32RR690 pKa = 3.28
Molecular weight: 75.82 kDa
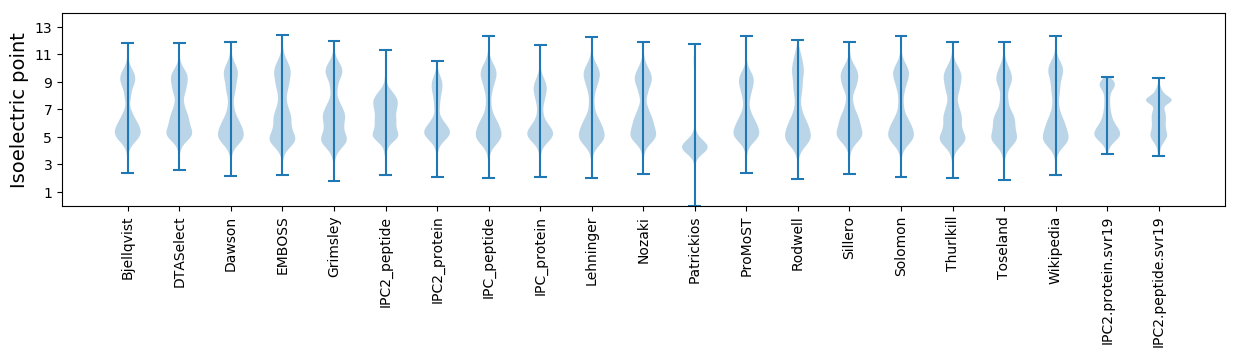
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7DNU0|A0A1G7DNU0_9FLAO Uncharacterized protein OS=Pricia antarctica OX=641691 GN=SAMN05421636_105406 PE=4 SV=1
MM1 pKa = 7.47LFVFDD6 pKa = 4.44ICPKK10 pKa = 10.42KK11 pKa = 10.67SQTPNRR17 pKa = 11.84AVWNLGFGTSIALGVIPLGLAATAVYY43 pKa = 9.4WFIVNKK49 pKa = 10.22NSSLFLVCLMRR60 pKa = 11.84CSMNSIASTGFISAKK75 pKa = 7.3YY76 pKa = 9.68LRR78 pKa = 11.84RR79 pKa = 11.84IHH81 pKa = 5.96MRR83 pKa = 3.27
MM1 pKa = 7.47LFVFDD6 pKa = 4.44ICPKK10 pKa = 10.42KK11 pKa = 10.67SQTPNRR17 pKa = 11.84AVWNLGFGTSIALGVIPLGLAATAVYY43 pKa = 9.4WFIVNKK49 pKa = 10.22NSSLFLVCLMRR60 pKa = 11.84CSMNSIASTGFISAKK75 pKa = 7.3YY76 pKa = 9.68LRR78 pKa = 11.84RR79 pKa = 11.84IHH81 pKa = 5.96MRR83 pKa = 3.27
Molecular weight: 9.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1420157 |
29 |
4883 |
340.4 |
38.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.924 ± 0.037 | 0.728 ± 0.013 |
5.972 ± 0.034 | 6.564 ± 0.029 |
5.041 ± 0.031 | 7.049 ± 0.039 |
1.896 ± 0.021 | 7.216 ± 0.03 |
7.168 ± 0.048 | 9.357 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.404 ± 0.02 | 5.36 ± 0.034 |
3.76 ± 0.025 | 3.355 ± 0.02 |
3.985 ± 0.025 | 6.248 ± 0.031 |
5.696 ± 0.039 | 6.264 ± 0.027 |
1.161 ± 0.014 | 3.851 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
