
Hubei sobemo-like virus 45
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
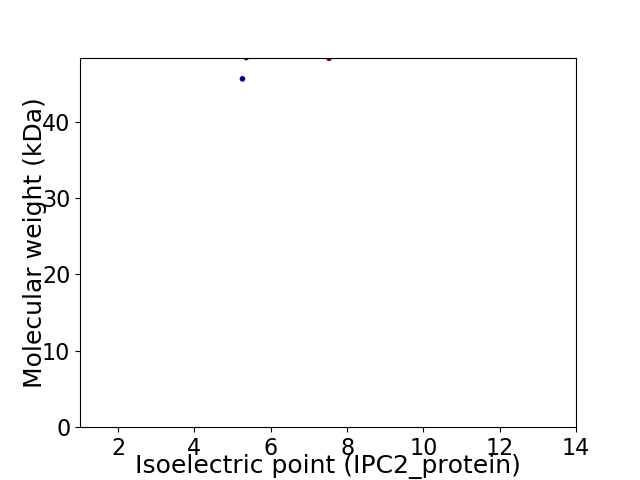
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEI0|A0A1L3KEI0_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 45 OX=1923233 PE=4 SV=1
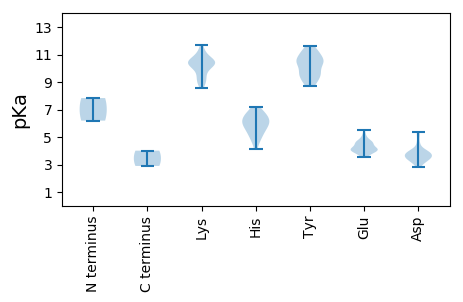
MM1 pKa = 6.19TQAMIPEE8 pKa = 5.11DD9 pKa = 3.86DD10 pKa = 4.03LRR12 pKa = 11.84EE13 pKa = 4.08KK14 pKa = 10.66VLSVMEE20 pKa = 4.59DD21 pKa = 2.87WYY23 pKa = 11.18KK24 pKa = 8.97PCRR27 pKa = 11.84WVLPDD32 pKa = 3.58DD33 pKa = 3.58WFSFEE38 pKa = 4.71RR39 pKa = 11.84FRR41 pKa = 11.84LVLNRR46 pKa = 11.84LDD48 pKa = 3.99RR49 pKa = 11.84SSSPGYY55 pKa = 8.91PYY57 pKa = 10.42CGYY60 pKa = 10.61KK61 pKa = 8.59PTIGEE66 pKa = 3.88WLGFDD71 pKa = 3.54GFVYY75 pKa = 10.64DD76 pKa = 5.06PIQVQLLWLDD86 pKa = 3.52VQRR89 pKa = 11.84VRR91 pKa = 11.84DD92 pKa = 4.04GEE94 pKa = 4.12SDD96 pKa = 3.34LVQRR100 pKa = 11.84VFIKK104 pKa = 10.63RR105 pKa = 11.84EE106 pKa = 3.62PHH108 pKa = 6.1KK109 pKa = 10.46KK110 pKa = 9.94SKK112 pKa = 10.22AAEE115 pKa = 4.23GRR117 pKa = 11.84WRR119 pKa = 11.84LIMAFPLDD127 pKa = 4.01HH128 pKa = 6.24QVFWHH133 pKa = 6.07MLFDD137 pKa = 3.87YY138 pKa = 11.24QNDD141 pKa = 4.0LEE143 pKa = 4.62ISHH146 pKa = 6.43ATSIPSQQGIWLHH159 pKa = 5.52GGAWRR164 pKa = 11.84GHH166 pKa = 4.16IQRR169 pKa = 11.84WKK171 pKa = 8.96QFGYY175 pKa = 10.88NVGLDD180 pKa = 3.33KK181 pKa = 11.27SAWDD185 pKa = 3.18WTYY188 pKa = 11.22PSWLLDD194 pKa = 2.85WDD196 pKa = 3.79LQFRR200 pKa = 11.84YY201 pKa = 10.48RR202 pKa = 11.84MGHH205 pKa = 4.84GSKK208 pKa = 9.43MEE210 pKa = 3.69EE211 pKa = 3.96WMNLASRR218 pKa = 11.84EE219 pKa = 4.03WKK221 pKa = 10.26LAFGVGAKK229 pKa = 10.39FITTQGWLLEE239 pKa = 4.14QQVPGVMKK247 pKa = 10.36SGAVVTIASNSHH259 pKa = 5.34AQAMLHH265 pKa = 6.04VLVCLDD271 pKa = 3.78EE272 pKa = 4.8GVDD275 pKa = 3.79YY276 pKa = 11.15EE277 pKa = 5.12PFPACCGDD285 pKa = 3.69DD286 pKa = 3.27TLQRR290 pKa = 11.84LDD292 pKa = 3.35QASVEE297 pKa = 4.62GYY299 pKa = 9.69AKK301 pKa = 10.55YY302 pKa = 10.65GVVVKK307 pKa = 10.47SASEE311 pKa = 3.76GLEE314 pKa = 3.91FMGHH318 pKa = 6.91DD319 pKa = 4.24FLDD322 pKa = 3.89TGPAPLYY329 pKa = 10.3LEE331 pKa = 4.08KK332 pKa = 10.76HH333 pKa = 5.85FSRR336 pKa = 11.84FLHH339 pKa = 6.73LEE341 pKa = 3.97EE342 pKa = 5.48EE343 pKa = 4.34IVPDD347 pKa = 5.35FIEE350 pKa = 4.56SMARR354 pKa = 11.84LYY356 pKa = 11.31VKK358 pKa = 9.63TPYY361 pKa = 9.62YY362 pKa = 9.42WLWEE366 pKa = 4.64HH367 pKa = 6.81ISVAMGCIVPSKK379 pKa = 10.12FALEE383 pKa = 3.58RR384 pKa = 11.84WYY386 pKa = 10.59DD387 pKa = 3.39HH388 pKa = 7.19SDD390 pKa = 2.9
MM1 pKa = 6.19TQAMIPEE8 pKa = 5.11DD9 pKa = 3.86DD10 pKa = 4.03LRR12 pKa = 11.84EE13 pKa = 4.08KK14 pKa = 10.66VLSVMEE20 pKa = 4.59DD21 pKa = 2.87WYY23 pKa = 11.18KK24 pKa = 8.97PCRR27 pKa = 11.84WVLPDD32 pKa = 3.58DD33 pKa = 3.58WFSFEE38 pKa = 4.71RR39 pKa = 11.84FRR41 pKa = 11.84LVLNRR46 pKa = 11.84LDD48 pKa = 3.99RR49 pKa = 11.84SSSPGYY55 pKa = 8.91PYY57 pKa = 10.42CGYY60 pKa = 10.61KK61 pKa = 8.59PTIGEE66 pKa = 3.88WLGFDD71 pKa = 3.54GFVYY75 pKa = 10.64DD76 pKa = 5.06PIQVQLLWLDD86 pKa = 3.52VQRR89 pKa = 11.84VRR91 pKa = 11.84DD92 pKa = 4.04GEE94 pKa = 4.12SDD96 pKa = 3.34LVQRR100 pKa = 11.84VFIKK104 pKa = 10.63RR105 pKa = 11.84EE106 pKa = 3.62PHH108 pKa = 6.1KK109 pKa = 10.46KK110 pKa = 9.94SKK112 pKa = 10.22AAEE115 pKa = 4.23GRR117 pKa = 11.84WRR119 pKa = 11.84LIMAFPLDD127 pKa = 4.01HH128 pKa = 6.24QVFWHH133 pKa = 6.07MLFDD137 pKa = 3.87YY138 pKa = 11.24QNDD141 pKa = 4.0LEE143 pKa = 4.62ISHH146 pKa = 6.43ATSIPSQQGIWLHH159 pKa = 5.52GGAWRR164 pKa = 11.84GHH166 pKa = 4.16IQRR169 pKa = 11.84WKK171 pKa = 8.96QFGYY175 pKa = 10.88NVGLDD180 pKa = 3.33KK181 pKa = 11.27SAWDD185 pKa = 3.18WTYY188 pKa = 11.22PSWLLDD194 pKa = 2.85WDD196 pKa = 3.79LQFRR200 pKa = 11.84YY201 pKa = 10.48RR202 pKa = 11.84MGHH205 pKa = 4.84GSKK208 pKa = 9.43MEE210 pKa = 3.69EE211 pKa = 3.96WMNLASRR218 pKa = 11.84EE219 pKa = 4.03WKK221 pKa = 10.26LAFGVGAKK229 pKa = 10.39FITTQGWLLEE239 pKa = 4.14QQVPGVMKK247 pKa = 10.36SGAVVTIASNSHH259 pKa = 5.34AQAMLHH265 pKa = 6.04VLVCLDD271 pKa = 3.78EE272 pKa = 4.8GVDD275 pKa = 3.79YY276 pKa = 11.15EE277 pKa = 5.12PFPACCGDD285 pKa = 3.69DD286 pKa = 3.27TLQRR290 pKa = 11.84LDD292 pKa = 3.35QASVEE297 pKa = 4.62GYY299 pKa = 9.69AKK301 pKa = 10.55YY302 pKa = 10.65GVVVKK307 pKa = 10.47SASEE311 pKa = 3.76GLEE314 pKa = 3.91FMGHH318 pKa = 6.91DD319 pKa = 4.24FLDD322 pKa = 3.89TGPAPLYY329 pKa = 10.3LEE331 pKa = 4.08KK332 pKa = 10.76HH333 pKa = 5.85FSRR336 pKa = 11.84FLHH339 pKa = 6.73LEE341 pKa = 3.97EE342 pKa = 5.48EE343 pKa = 4.34IVPDD347 pKa = 5.35FIEE350 pKa = 4.56SMARR354 pKa = 11.84LYY356 pKa = 11.31VKK358 pKa = 9.63TPYY361 pKa = 9.62YY362 pKa = 9.42WLWEE366 pKa = 4.64HH367 pKa = 6.81ISVAMGCIVPSKK379 pKa = 10.12FALEE383 pKa = 3.58RR384 pKa = 11.84WYY386 pKa = 10.59DD387 pKa = 3.39HH388 pKa = 7.19SDD390 pKa = 2.9
Molecular weight: 45.59 kDa
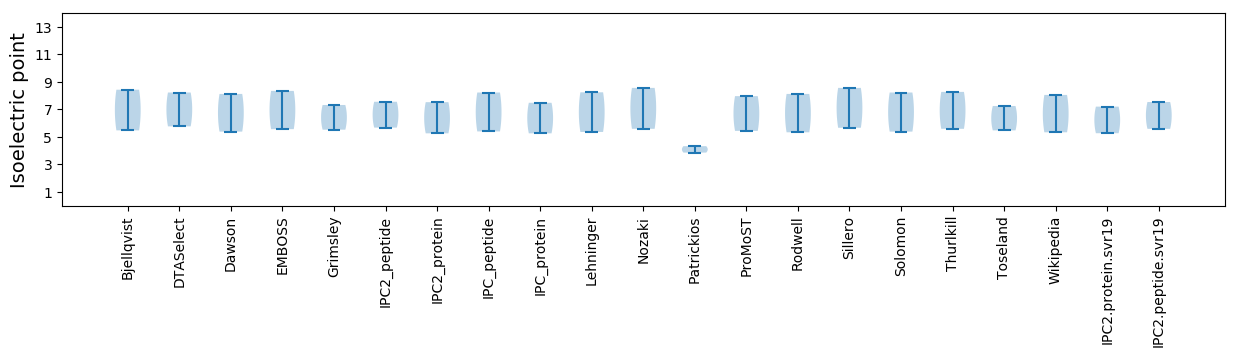
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEI0|A0A1L3KEI0_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 45 OX=1923233 PE=4 SV=1
MM1 pKa = 7.83PFFEE5 pKa = 4.8EE6 pKa = 4.58LVCFGGHH13 pKa = 5.57QLWNQPTTVWGRR25 pKa = 11.84LFDD28 pKa = 3.84CSAILGGSPGPFRR41 pKa = 11.84VAAYY45 pKa = 9.93CGAAGALAAGAYY57 pKa = 10.02VGFPKK62 pKa = 10.46LVNSVRR68 pKa = 11.84RR69 pKa = 11.84SFGWRR74 pKa = 11.84PPMKK78 pKa = 9.33MDD80 pKa = 3.52VSPASVTNHH89 pKa = 4.74GRR91 pKa = 11.84LAEE94 pKa = 4.09SRR96 pKa = 11.84FAGSAEE102 pKa = 4.2TAMSMAVGQVAVGLYY117 pKa = 8.92MEE119 pKa = 4.9GSFRR123 pKa = 11.84VSGSGFRR130 pKa = 11.84VDD132 pKa = 4.53VGDD135 pKa = 4.64LEE137 pKa = 4.43TLIVPAHH144 pKa = 4.93VWEE147 pKa = 4.32EE148 pKa = 4.04VVARR152 pKa = 11.84SGDD155 pKa = 3.5RR156 pKa = 11.84VVVRR160 pKa = 11.84GKK162 pKa = 10.49KK163 pKa = 10.17ADD165 pKa = 3.61VEE167 pKa = 4.27LLDD170 pKa = 3.88SEE172 pKa = 4.58VRR174 pKa = 11.84KK175 pKa = 10.28HH176 pKa = 6.47EE177 pKa = 3.58IHH179 pKa = 6.46TDD181 pKa = 3.0VFLVVMPPNAFSKK194 pKa = 10.78AGVGRR199 pKa = 11.84STIVPIVGKK208 pKa = 10.49LSAKK212 pKa = 9.74VAGPHH217 pKa = 5.93GMGTFGTLSLGEE229 pKa = 3.91RR230 pKa = 11.84AAFGTTWYY238 pKa = 10.47HH239 pKa = 5.38GTTLPGYY246 pKa = 9.35SGSPYY251 pKa = 9.42TVSGNIAAMHH261 pKa = 5.87LRR263 pKa = 11.84GAEE266 pKa = 4.11VQHH269 pKa = 6.37GPNIGVSAQMLYY281 pKa = 8.97VTAKK285 pKa = 10.04HH286 pKa = 5.94VLNQRR291 pKa = 11.84LEE293 pKa = 4.13EE294 pKa = 4.17TYY296 pKa = 10.7EE297 pKa = 3.94WLTEE301 pKa = 3.88MAQRR305 pKa = 11.84KK306 pKa = 9.21ADD308 pKa = 3.51IEE310 pKa = 4.21YY311 pKa = 9.51DD312 pKa = 3.71QSWRR316 pKa = 11.84SQDD319 pKa = 3.29TVRR322 pKa = 11.84VKK324 pKa = 11.06VKK326 pKa = 10.28GQFAIVDD333 pKa = 3.69KK334 pKa = 11.17DD335 pKa = 3.78AMNSAYY341 pKa = 10.37GDD343 pKa = 3.44DD344 pKa = 3.14WHH346 pKa = 6.66NQVRR350 pKa = 11.84YY351 pKa = 9.7RR352 pKa = 11.84DD353 pKa = 3.66YY354 pKa = 11.61TEE356 pKa = 4.02DD357 pKa = 3.36SKK359 pKa = 11.67RR360 pKa = 11.84NRR362 pKa = 11.84HH363 pKa = 5.33SALTQEE369 pKa = 4.56SSGNAARR376 pKa = 11.84PGVLPSPVGPQGLVAYY392 pKa = 9.51AGPSCQNTGSVSCEE406 pKa = 3.99EE407 pKa = 4.32LCMGLTDD414 pKa = 5.27AEE416 pKa = 4.57LKK418 pKa = 10.52RR419 pKa = 11.84LSSVANRR426 pKa = 11.84YY427 pKa = 8.69YY428 pKa = 10.31SQRR431 pKa = 11.84LNALKK436 pKa = 8.97ITSGQGQTPTII447 pKa = 4.01
MM1 pKa = 7.83PFFEE5 pKa = 4.8EE6 pKa = 4.58LVCFGGHH13 pKa = 5.57QLWNQPTTVWGRR25 pKa = 11.84LFDD28 pKa = 3.84CSAILGGSPGPFRR41 pKa = 11.84VAAYY45 pKa = 9.93CGAAGALAAGAYY57 pKa = 10.02VGFPKK62 pKa = 10.46LVNSVRR68 pKa = 11.84RR69 pKa = 11.84SFGWRR74 pKa = 11.84PPMKK78 pKa = 9.33MDD80 pKa = 3.52VSPASVTNHH89 pKa = 4.74GRR91 pKa = 11.84LAEE94 pKa = 4.09SRR96 pKa = 11.84FAGSAEE102 pKa = 4.2TAMSMAVGQVAVGLYY117 pKa = 8.92MEE119 pKa = 4.9GSFRR123 pKa = 11.84VSGSGFRR130 pKa = 11.84VDD132 pKa = 4.53VGDD135 pKa = 4.64LEE137 pKa = 4.43TLIVPAHH144 pKa = 4.93VWEE147 pKa = 4.32EE148 pKa = 4.04VVARR152 pKa = 11.84SGDD155 pKa = 3.5RR156 pKa = 11.84VVVRR160 pKa = 11.84GKK162 pKa = 10.49KK163 pKa = 10.17ADD165 pKa = 3.61VEE167 pKa = 4.27LLDD170 pKa = 3.88SEE172 pKa = 4.58VRR174 pKa = 11.84KK175 pKa = 10.28HH176 pKa = 6.47EE177 pKa = 3.58IHH179 pKa = 6.46TDD181 pKa = 3.0VFLVVMPPNAFSKK194 pKa = 10.78AGVGRR199 pKa = 11.84STIVPIVGKK208 pKa = 10.49LSAKK212 pKa = 9.74VAGPHH217 pKa = 5.93GMGTFGTLSLGEE229 pKa = 3.91RR230 pKa = 11.84AAFGTTWYY238 pKa = 10.47HH239 pKa = 5.38GTTLPGYY246 pKa = 9.35SGSPYY251 pKa = 9.42TVSGNIAAMHH261 pKa = 5.87LRR263 pKa = 11.84GAEE266 pKa = 4.11VQHH269 pKa = 6.37GPNIGVSAQMLYY281 pKa = 8.97VTAKK285 pKa = 10.04HH286 pKa = 5.94VLNQRR291 pKa = 11.84LEE293 pKa = 4.13EE294 pKa = 4.17TYY296 pKa = 10.7EE297 pKa = 3.94WLTEE301 pKa = 3.88MAQRR305 pKa = 11.84KK306 pKa = 9.21ADD308 pKa = 3.51IEE310 pKa = 4.21YY311 pKa = 9.51DD312 pKa = 3.71QSWRR316 pKa = 11.84SQDD319 pKa = 3.29TVRR322 pKa = 11.84VKK324 pKa = 11.06VKK326 pKa = 10.28GQFAIVDD333 pKa = 3.69KK334 pKa = 11.17DD335 pKa = 3.78AMNSAYY341 pKa = 10.37GDD343 pKa = 3.44DD344 pKa = 3.14WHH346 pKa = 6.66NQVRR350 pKa = 11.84YY351 pKa = 9.7RR352 pKa = 11.84DD353 pKa = 3.66YY354 pKa = 11.61TEE356 pKa = 4.02DD357 pKa = 3.36SKK359 pKa = 11.67RR360 pKa = 11.84NRR362 pKa = 11.84HH363 pKa = 5.33SALTQEE369 pKa = 4.56SSGNAARR376 pKa = 11.84PGVLPSPVGPQGLVAYY392 pKa = 9.51AGPSCQNTGSVSCEE406 pKa = 3.99EE407 pKa = 4.32LCMGLTDD414 pKa = 5.27AEE416 pKa = 4.57LKK418 pKa = 10.52RR419 pKa = 11.84LSSVANRR426 pKa = 11.84YY427 pKa = 8.69YY428 pKa = 10.31SQRR431 pKa = 11.84LNALKK436 pKa = 8.97ITSGQGQTPTII447 pKa = 4.01
Molecular weight: 48.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
837 |
390 |
447 |
418.5 |
46.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.766 ± 1.426 | 1.434 ± 0.07 |
5.496 ± 1.13 | 5.735 ± 0.453 |
4.182 ± 0.635 | 9.08 ± 1.275 |
3.106 ± 0.324 | 3.106 ± 0.496 |
4.062 ± 0.371 | 8.244 ± 1.006 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.106 ± 0.152 | 2.27 ± 0.663 |
4.898 ± 0.018 | 4.182 ± 0.291 |
5.854 ± 0.315 | 7.407 ± 0.669 |
4.182 ± 1.085 | 8.722 ± 1.035 |
3.345 ± 1.196 | 3.823 ± 0.36 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
