
Azospirillum sp. CAG:260
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Azospirillaceae; Azospirillum; environmental samples
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
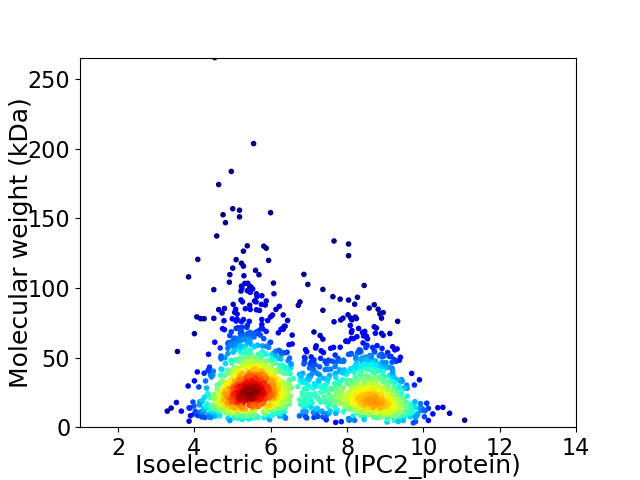
Virtual 2D-PAGE plot for 1762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6HQV3|R6HQV3_9PROT Shikimate kinase OS=Azospirillum sp. CAG:260 OX=1262706 GN=aroK PE=3 SV=1
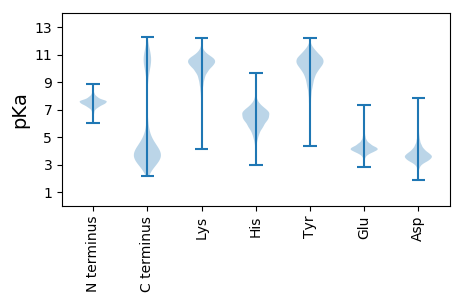
MM1 pKa = 7.41TSYY4 pKa = 10.89ILQVHH9 pKa = 5.71SRR11 pKa = 11.84YY12 pKa = 10.05LDD14 pKa = 3.73LPLVPPSTGIARR26 pKa = 11.84HH27 pKa = 5.34TGYY30 pKa = 11.17DD31 pKa = 3.99LVTLNDD37 pKa = 3.4NGEE40 pKa = 3.92PSFINSVDD48 pKa = 3.16LTTRR52 pKa = 11.84NGRR55 pKa = 11.84IYY57 pKa = 9.79MVFVEE62 pKa = 5.39GSDD65 pKa = 3.53GQPFIVNVSGEE76 pKa = 4.03KK77 pKa = 10.34QIWPKK82 pKa = 10.11KK83 pKa = 7.88IHH85 pKa = 6.3IGEE88 pKa = 4.17VTADD92 pKa = 3.37NEE94 pKa = 4.72GPNFTVTQEE103 pKa = 3.99GAEE106 pKa = 4.12KK107 pKa = 10.88YY108 pKa = 10.07DD109 pKa = 3.68FKK111 pKa = 10.91TATVLTGDD119 pKa = 3.78VAKK122 pKa = 10.58HH123 pKa = 4.57VFDD126 pKa = 5.54WIVFDD131 pKa = 4.18AKK133 pKa = 10.76YY134 pKa = 10.72RR135 pKa = 11.84NALSMVSNGLTDD147 pKa = 3.87YY148 pKa = 11.25DD149 pKa = 4.41VNAPNCNTWTNYY161 pKa = 9.65IDD163 pKa = 3.33LTYY166 pKa = 10.62IKK168 pKa = 10.23GSQSVFDD175 pKa = 4.12KK176 pKa = 11.35LGGGWYY182 pKa = 9.23PGKK185 pKa = 10.39DD186 pKa = 2.82KK187 pKa = 11.07TFINSTSGDD196 pKa = 3.75DD197 pKa = 5.23LRR199 pKa = 11.84DD200 pKa = 3.54CKK202 pKa = 10.57IIEE205 pKa = 4.84DD206 pKa = 3.54ISRR209 pKa = 11.84KK210 pKa = 9.11FFEE213 pKa = 5.58LYY215 pKa = 10.48SADD218 pKa = 3.72FNFGNLSITLEE229 pKa = 4.21DD230 pKa = 3.26VKK232 pKa = 10.92HH233 pKa = 5.9YY234 pKa = 10.73SNTMDD239 pKa = 3.39PVGFVTRR246 pKa = 11.84ISDD249 pKa = 3.73GVRR252 pKa = 11.84SYY254 pKa = 11.62LFDD257 pKa = 4.45CEE259 pKa = 4.5VVNSYY264 pKa = 10.3VQDD267 pKa = 3.12GHH269 pKa = 6.67YY270 pKa = 10.85FEE272 pKa = 6.7DD273 pKa = 3.81NTPGGSYY280 pKa = 9.83IVAGKK285 pKa = 9.69GASEE289 pKa = 4.19TIIAGDD295 pKa = 3.71GDD297 pKa = 4.77DD298 pKa = 5.54IIDD301 pKa = 3.83AGAGEE306 pKa = 5.06GEE308 pKa = 4.34TAVAKK313 pKa = 9.95YY314 pKa = 10.71VNLGGGDD321 pKa = 3.51NLYY324 pKa = 11.05VGGAAADD331 pKa = 3.92EE332 pKa = 4.85VNTGTDD338 pKa = 2.96NSGSVANEE346 pKa = 3.34NNVNHH351 pKa = 5.89VFLGAGSDD359 pKa = 3.34TFYY362 pKa = 10.94GGKK365 pKa = 10.09GADD368 pKa = 3.35IVDD371 pKa = 4.26GGSGDD376 pKa = 3.93LTRR379 pKa = 11.84LQALPQFIDD388 pKa = 3.32SGVTTEE394 pKa = 5.96SMADD398 pKa = 3.57DD399 pKa = 3.91EE400 pKa = 4.76NAVNRR405 pKa = 11.84INLGVGNDD413 pKa = 3.42VYY415 pKa = 11.18FGGKK419 pKa = 9.92GKK421 pKa = 10.27DD422 pKa = 3.09AVYY425 pKa = 10.56GGDD428 pKa = 4.03GDD430 pKa = 4.96DD431 pKa = 4.19IIYY434 pKa = 10.85GGDD437 pKa = 3.56CPGGFDD443 pKa = 5.46GRR445 pKa = 11.84NEE447 pKa = 4.27LYY449 pKa = 11.03GEE451 pKa = 4.54AGDD454 pKa = 5.93DD455 pKa = 4.13YY456 pKa = 11.24ICGGRR461 pKa = 11.84SDD463 pKa = 5.14DD464 pKa = 4.21VISGGEE470 pKa = 4.21GNDD473 pKa = 3.3VIYY476 pKa = 10.88GNGGNDD482 pKa = 3.25VLMADD487 pKa = 4.06AGSDD491 pKa = 3.27IIYY494 pKa = 10.71AGVGYY499 pKa = 10.85NNIYY503 pKa = 10.55CGEE506 pKa = 4.06NDD508 pKa = 3.37SCRR511 pKa = 11.84DD512 pKa = 3.31IVVLNNDD519 pKa = 2.86VNGRR523 pKa = 11.84DD524 pKa = 3.87DD525 pKa = 3.72IYY527 pKa = 11.2NITSQDD533 pKa = 3.47IICCVGGFDD542 pKa = 4.55FSRR545 pKa = 11.84SLGYY549 pKa = 9.98YY550 pKa = 9.45SPLWGVKK557 pKa = 9.76GNSINFQTTGDD568 pKa = 4.22NVPLIYY574 pKa = 10.81DD575 pKa = 3.69ADD577 pKa = 3.7GKK579 pKa = 9.3VYY581 pKa = 10.17RR582 pKa = 11.84WRR584 pKa = 11.84EE585 pKa = 3.65ACTDD589 pKa = 3.43ADD591 pKa = 4.84GITHH595 pKa = 6.77SSQYY599 pKa = 9.56IDD601 pKa = 3.03TGIRR605 pKa = 11.84YY606 pKa = 7.45PAYY609 pKa = 9.98GDD611 pKa = 3.94GEE613 pKa = 4.3PEE615 pKa = 3.93EE616 pKa = 4.97EE617 pKa = 4.44DD618 pKa = 3.9LKK620 pKa = 11.51KK621 pKa = 10.86LLSAA625 pKa = 5.11
MM1 pKa = 7.41TSYY4 pKa = 10.89ILQVHH9 pKa = 5.71SRR11 pKa = 11.84YY12 pKa = 10.05LDD14 pKa = 3.73LPLVPPSTGIARR26 pKa = 11.84HH27 pKa = 5.34TGYY30 pKa = 11.17DD31 pKa = 3.99LVTLNDD37 pKa = 3.4NGEE40 pKa = 3.92PSFINSVDD48 pKa = 3.16LTTRR52 pKa = 11.84NGRR55 pKa = 11.84IYY57 pKa = 9.79MVFVEE62 pKa = 5.39GSDD65 pKa = 3.53GQPFIVNVSGEE76 pKa = 4.03KK77 pKa = 10.34QIWPKK82 pKa = 10.11KK83 pKa = 7.88IHH85 pKa = 6.3IGEE88 pKa = 4.17VTADD92 pKa = 3.37NEE94 pKa = 4.72GPNFTVTQEE103 pKa = 3.99GAEE106 pKa = 4.12KK107 pKa = 10.88YY108 pKa = 10.07DD109 pKa = 3.68FKK111 pKa = 10.91TATVLTGDD119 pKa = 3.78VAKK122 pKa = 10.58HH123 pKa = 4.57VFDD126 pKa = 5.54WIVFDD131 pKa = 4.18AKK133 pKa = 10.76YY134 pKa = 10.72RR135 pKa = 11.84NALSMVSNGLTDD147 pKa = 3.87YY148 pKa = 11.25DD149 pKa = 4.41VNAPNCNTWTNYY161 pKa = 9.65IDD163 pKa = 3.33LTYY166 pKa = 10.62IKK168 pKa = 10.23GSQSVFDD175 pKa = 4.12KK176 pKa = 11.35LGGGWYY182 pKa = 9.23PGKK185 pKa = 10.39DD186 pKa = 2.82KK187 pKa = 11.07TFINSTSGDD196 pKa = 3.75DD197 pKa = 5.23LRR199 pKa = 11.84DD200 pKa = 3.54CKK202 pKa = 10.57IIEE205 pKa = 4.84DD206 pKa = 3.54ISRR209 pKa = 11.84KK210 pKa = 9.11FFEE213 pKa = 5.58LYY215 pKa = 10.48SADD218 pKa = 3.72FNFGNLSITLEE229 pKa = 4.21DD230 pKa = 3.26VKK232 pKa = 10.92HH233 pKa = 5.9YY234 pKa = 10.73SNTMDD239 pKa = 3.39PVGFVTRR246 pKa = 11.84ISDD249 pKa = 3.73GVRR252 pKa = 11.84SYY254 pKa = 11.62LFDD257 pKa = 4.45CEE259 pKa = 4.5VVNSYY264 pKa = 10.3VQDD267 pKa = 3.12GHH269 pKa = 6.67YY270 pKa = 10.85FEE272 pKa = 6.7DD273 pKa = 3.81NTPGGSYY280 pKa = 9.83IVAGKK285 pKa = 9.69GASEE289 pKa = 4.19TIIAGDD295 pKa = 3.71GDD297 pKa = 4.77DD298 pKa = 5.54IIDD301 pKa = 3.83AGAGEE306 pKa = 5.06GEE308 pKa = 4.34TAVAKK313 pKa = 9.95YY314 pKa = 10.71VNLGGGDD321 pKa = 3.51NLYY324 pKa = 11.05VGGAAADD331 pKa = 3.92EE332 pKa = 4.85VNTGTDD338 pKa = 2.96NSGSVANEE346 pKa = 3.34NNVNHH351 pKa = 5.89VFLGAGSDD359 pKa = 3.34TFYY362 pKa = 10.94GGKK365 pKa = 10.09GADD368 pKa = 3.35IVDD371 pKa = 4.26GGSGDD376 pKa = 3.93LTRR379 pKa = 11.84LQALPQFIDD388 pKa = 3.32SGVTTEE394 pKa = 5.96SMADD398 pKa = 3.57DD399 pKa = 3.91EE400 pKa = 4.76NAVNRR405 pKa = 11.84INLGVGNDD413 pKa = 3.42VYY415 pKa = 11.18FGGKK419 pKa = 9.92GKK421 pKa = 10.27DD422 pKa = 3.09AVYY425 pKa = 10.56GGDD428 pKa = 4.03GDD430 pKa = 4.96DD431 pKa = 4.19IIYY434 pKa = 10.85GGDD437 pKa = 3.56CPGGFDD443 pKa = 5.46GRR445 pKa = 11.84NEE447 pKa = 4.27LYY449 pKa = 11.03GEE451 pKa = 4.54AGDD454 pKa = 5.93DD455 pKa = 4.13YY456 pKa = 11.24ICGGRR461 pKa = 11.84SDD463 pKa = 5.14DD464 pKa = 4.21VISGGEE470 pKa = 4.21GNDD473 pKa = 3.3VIYY476 pKa = 10.88GNGGNDD482 pKa = 3.25VLMADD487 pKa = 4.06AGSDD491 pKa = 3.27IIYY494 pKa = 10.71AGVGYY499 pKa = 10.85NNIYY503 pKa = 10.55CGEE506 pKa = 4.06NDD508 pKa = 3.37SCRR511 pKa = 11.84DD512 pKa = 3.31IVVLNNDD519 pKa = 2.86VNGRR523 pKa = 11.84DD524 pKa = 3.87DD525 pKa = 3.72IYY527 pKa = 11.2NITSQDD533 pKa = 3.47IICCVGGFDD542 pKa = 4.55FSRR545 pKa = 11.84SLGYY549 pKa = 9.98YY550 pKa = 9.45SPLWGVKK557 pKa = 9.76GNSINFQTTGDD568 pKa = 4.22NVPLIYY574 pKa = 10.81DD575 pKa = 3.69ADD577 pKa = 3.7GKK579 pKa = 9.3VYY581 pKa = 10.17RR582 pKa = 11.84WRR584 pKa = 11.84EE585 pKa = 3.65ACTDD589 pKa = 3.43ADD591 pKa = 4.84GITHH595 pKa = 6.77SSQYY599 pKa = 9.56IDD601 pKa = 3.03TGIRR605 pKa = 11.84YY606 pKa = 7.45PAYY609 pKa = 9.98GDD611 pKa = 3.94GEE613 pKa = 4.3PEE615 pKa = 3.93EE616 pKa = 4.97EE617 pKa = 4.44DD618 pKa = 3.9LKK620 pKa = 11.51KK621 pKa = 10.86LLSAA625 pKa = 5.11
Molecular weight: 67.21 kDa
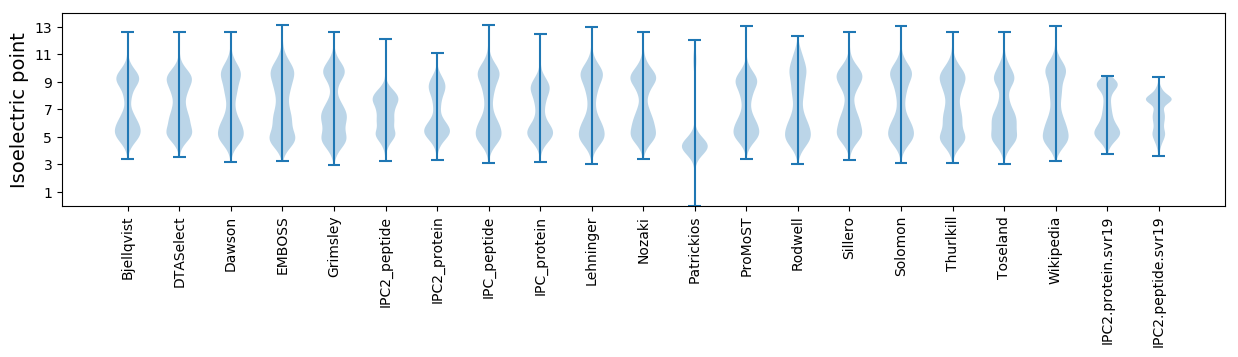
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6I3Z9|R6I3Z9_9PROT Ribonuclease H OS=Azospirillum sp. CAG:260 OX=1262706 GN=rnhA PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 8.92VLAARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.95HH37 pKa = 5.78GRR39 pKa = 11.84AKK41 pKa = 10.77LSAA44 pKa = 3.95
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 8.92VLAARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.95HH37 pKa = 5.78GRR39 pKa = 11.84AKK41 pKa = 10.77LSAA44 pKa = 3.95
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
526901 |
29 |
2605 |
299.0 |
33.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.57 ± 0.065 | 1.235 ± 0.03 |
5.644 ± 0.042 | 6.815 ± 0.071 |
4.326 ± 0.047 | 6.668 ± 0.077 |
1.627 ± 0.025 | 6.755 ± 0.055 |
7.01 ± 0.067 | 9.506 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.54 ± 0.026 | 4.981 ± 0.055 |
3.869 ± 0.042 | 3.486 ± 0.041 |
4.987 ± 0.048 | 5.842 ± 0.051 |
4.881 ± 0.047 | 6.55 ± 0.056 |
1.056 ± 0.022 | 3.65 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
