
Diaporthe ambigua RNA virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.93
Get precalculated fractions of proteins
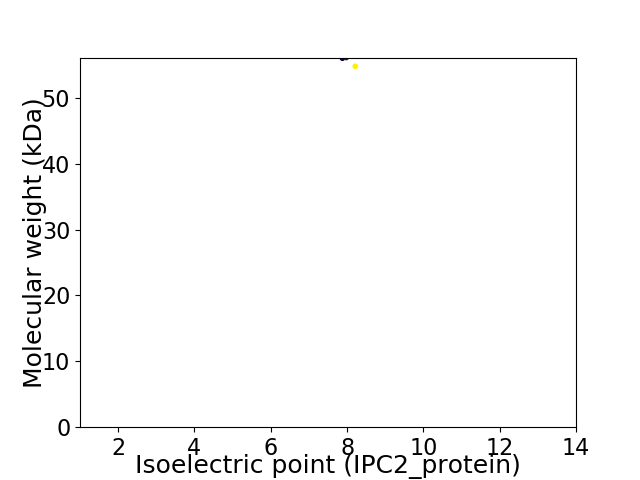
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9Q9R2|Q9Q9R2_9VIRU ORF1 OS=Diaporthe ambigua RNA virus 1 OX=111470 PE=4 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84FFNEE6 pKa = 4.41LMMTTDD12 pKa = 2.54IATYY16 pKa = 10.21YY17 pKa = 10.97SFASWYY23 pKa = 10.36GRR25 pKa = 11.84TFGYY29 pKa = 10.22GVPFWFIWRR38 pKa = 11.84DD39 pKa = 3.27FTLYY43 pKa = 10.8VGFDD47 pKa = 3.46VPEE50 pKa = 4.09EE51 pKa = 3.9LVAYY55 pKa = 9.66ALSLFSIAWLTTLIYY70 pKa = 9.86FHH72 pKa = 6.95LWATAFVAVAWFLGVVMLVRR92 pKa = 11.84AILRR96 pKa = 11.84FVEE99 pKa = 4.36WFTFGLAPFVGGFVSSSTWVVSCLCRR125 pKa = 11.84ALMYY129 pKa = 10.59LFFYY133 pKa = 10.63LPPPLVAGQLLKK145 pKa = 10.31LTLSLLWSLVWEE157 pKa = 4.94SIWLPVDD164 pKa = 3.78LWHH167 pKa = 7.11LGVGSWRR174 pKa = 11.84LGSVGWDD181 pKa = 3.12GSNRR185 pKa = 11.84VVQLGGFCFRR195 pKa = 11.84IRR197 pKa = 11.84FSDD200 pKa = 3.7FVYY203 pKa = 9.8PYY205 pKa = 10.26RR206 pKa = 11.84VLVLPGFVLGVLHH219 pKa = 6.16VLPGGFWLCFPSFKK233 pKa = 10.67FEE235 pKa = 3.94QLGVLGSLFIYY246 pKa = 10.11PLVEE250 pKa = 3.84TKK252 pKa = 10.78LIFVLGWVDD261 pKa = 3.49VAFVLPWLAAVLVGVSFSYY280 pKa = 10.91RR281 pKa = 11.84EE282 pKa = 4.14VPFSRR287 pKa = 11.84ADD289 pKa = 3.41LQGYY293 pKa = 9.51LGLSEE298 pKa = 4.83PSQDD302 pKa = 3.03ASVLSEE308 pKa = 4.57LDD310 pKa = 3.38ALLKK314 pKa = 10.79AKK316 pKa = 10.46SFADD320 pKa = 3.54AEE322 pKa = 4.67GVALVAAVGARR333 pKa = 11.84GGVPRR338 pKa = 11.84LRR340 pKa = 11.84ARR342 pKa = 11.84GKK344 pKa = 9.56LACRR348 pKa = 11.84LQALLGGRR356 pKa = 11.84KK357 pKa = 9.23GCVGAFFSGRR367 pKa = 11.84WVPDD371 pKa = 3.46LPASDD376 pKa = 4.77RR377 pKa = 11.84SPALNAILAVQRR389 pKa = 11.84EE390 pKa = 4.27QVKK393 pKa = 10.3VLGVGTFPTKK403 pKa = 10.56SGVGGDD409 pKa = 3.79YY410 pKa = 11.22VLVEE414 pKa = 4.32SRR416 pKa = 11.84SGDD419 pKa = 3.1RR420 pKa = 11.84RR421 pKa = 11.84VILPSLLSRR430 pKa = 11.84LCLYY434 pKa = 10.77SSLRR438 pKa = 11.84EE439 pKa = 3.84RR440 pKa = 11.84DD441 pKa = 3.29EE442 pKa = 4.1KK443 pKa = 11.45LLVGLRR449 pKa = 11.84SRR451 pKa = 11.84AVEE454 pKa = 3.85WCRR457 pKa = 11.84LEE459 pKa = 4.98QVPDD463 pKa = 3.87WVPLLCLSSSVADD476 pKa = 3.48AAEE479 pKa = 3.87ISGPEE484 pKa = 3.62VEE486 pKa = 4.0ARR488 pKa = 11.84RR489 pKa = 11.84RR490 pKa = 11.84LRR492 pKa = 11.84SSLGIDD498 pKa = 3.27TLASFF503 pKa = 5.05
MM1 pKa = 7.82RR2 pKa = 11.84FFNEE6 pKa = 4.41LMMTTDD12 pKa = 2.54IATYY16 pKa = 10.21YY17 pKa = 10.97SFASWYY23 pKa = 10.36GRR25 pKa = 11.84TFGYY29 pKa = 10.22GVPFWFIWRR38 pKa = 11.84DD39 pKa = 3.27FTLYY43 pKa = 10.8VGFDD47 pKa = 3.46VPEE50 pKa = 4.09EE51 pKa = 3.9LVAYY55 pKa = 9.66ALSLFSIAWLTTLIYY70 pKa = 9.86FHH72 pKa = 6.95LWATAFVAVAWFLGVVMLVRR92 pKa = 11.84AILRR96 pKa = 11.84FVEE99 pKa = 4.36WFTFGLAPFVGGFVSSSTWVVSCLCRR125 pKa = 11.84ALMYY129 pKa = 10.59LFFYY133 pKa = 10.63LPPPLVAGQLLKK145 pKa = 10.31LTLSLLWSLVWEE157 pKa = 4.94SIWLPVDD164 pKa = 3.78LWHH167 pKa = 7.11LGVGSWRR174 pKa = 11.84LGSVGWDD181 pKa = 3.12GSNRR185 pKa = 11.84VVQLGGFCFRR195 pKa = 11.84IRR197 pKa = 11.84FSDD200 pKa = 3.7FVYY203 pKa = 9.8PYY205 pKa = 10.26RR206 pKa = 11.84VLVLPGFVLGVLHH219 pKa = 6.16VLPGGFWLCFPSFKK233 pKa = 10.67FEE235 pKa = 3.94QLGVLGSLFIYY246 pKa = 10.11PLVEE250 pKa = 3.84TKK252 pKa = 10.78LIFVLGWVDD261 pKa = 3.49VAFVLPWLAAVLVGVSFSYY280 pKa = 10.91RR281 pKa = 11.84EE282 pKa = 4.14VPFSRR287 pKa = 11.84ADD289 pKa = 3.41LQGYY293 pKa = 9.51LGLSEE298 pKa = 4.83PSQDD302 pKa = 3.03ASVLSEE308 pKa = 4.57LDD310 pKa = 3.38ALLKK314 pKa = 10.79AKK316 pKa = 10.46SFADD320 pKa = 3.54AEE322 pKa = 4.67GVALVAAVGARR333 pKa = 11.84GGVPRR338 pKa = 11.84LRR340 pKa = 11.84ARR342 pKa = 11.84GKK344 pKa = 9.56LACRR348 pKa = 11.84LQALLGGRR356 pKa = 11.84KK357 pKa = 9.23GCVGAFFSGRR367 pKa = 11.84WVPDD371 pKa = 3.46LPASDD376 pKa = 4.77RR377 pKa = 11.84SPALNAILAVQRR389 pKa = 11.84EE390 pKa = 4.27QVKK393 pKa = 10.3VLGVGTFPTKK403 pKa = 10.56SGVGGDD409 pKa = 3.79YY410 pKa = 11.22VLVEE414 pKa = 4.32SRR416 pKa = 11.84SGDD419 pKa = 3.1RR420 pKa = 11.84RR421 pKa = 11.84VILPSLLSRR430 pKa = 11.84LCLYY434 pKa = 10.77SSLRR438 pKa = 11.84EE439 pKa = 3.84RR440 pKa = 11.84DD441 pKa = 3.29EE442 pKa = 4.1KK443 pKa = 11.45LLVGLRR449 pKa = 11.84SRR451 pKa = 11.84AVEE454 pKa = 3.85WCRR457 pKa = 11.84LEE459 pKa = 4.98QVPDD463 pKa = 3.87WVPLLCLSSSVADD476 pKa = 3.48AAEE479 pKa = 3.87ISGPEE484 pKa = 3.62VEE486 pKa = 4.0ARR488 pKa = 11.84RR489 pKa = 11.84RR490 pKa = 11.84LRR492 pKa = 11.84SSLGIDD498 pKa = 3.27TLASFF503 pKa = 5.05
Molecular weight: 56.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9Q9R2|Q9Q9R2_9VIRU ORF1 OS=Diaporthe ambigua RNA virus 1 OX=111470 PE=4 SV=1
MM1 pKa = 7.35FVPFPLPCEE10 pKa = 4.47GTWLPQVHH18 pKa = 5.87RR19 pKa = 11.84VCPHH23 pKa = 6.02NEE25 pKa = 3.31VGALCRR31 pKa = 11.84RR32 pKa = 11.84VLAPLPAQVQEE43 pKa = 4.24RR44 pKa = 11.84VSPGFSRR51 pKa = 11.84AMGLAVGLARR61 pKa = 11.84FYY63 pKa = 9.24TQPRR67 pKa = 11.84WSLEE71 pKa = 3.97RR72 pKa = 11.84TALSYY77 pKa = 11.27DD78 pKa = 3.1GALRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 9.66LGAMDD90 pKa = 5.96SLLDD94 pKa = 3.51WPLDD98 pKa = 3.52VGKK101 pKa = 10.17DD102 pKa = 3.4SRR104 pKa = 11.84LDD106 pKa = 3.53CFLKK110 pKa = 10.84AEE112 pKa = 4.52KK113 pKa = 10.76LNTLIKK119 pKa = 10.16RR120 pKa = 11.84PKK122 pKa = 9.0PRR124 pKa = 11.84LIFPRR129 pKa = 11.84SPRR132 pKa = 11.84YY133 pKa = 9.95NLVLASRR140 pKa = 11.84LKK142 pKa = 10.71PFEE145 pKa = 3.91HH146 pKa = 6.89WLWSHH151 pKa = 6.11LTAEE155 pKa = 4.54RR156 pKa = 11.84LGTSGRR162 pKa = 11.84GRR164 pKa = 11.84YY165 pKa = 7.4VAKK168 pKa = 10.39GLNLSQRR175 pKa = 11.84ARR177 pKa = 11.84LIVKK181 pKa = 9.97KK182 pKa = 9.47MAGFSRR188 pKa = 11.84CAVLEE193 pKa = 3.99VDD195 pKa = 4.33GKK197 pKa = 11.02AFEE200 pKa = 4.27AHH202 pKa = 6.07VGPSSIEE209 pKa = 3.81QEE211 pKa = 3.88HH212 pKa = 5.84RR213 pKa = 11.84VYY215 pKa = 10.66KK216 pKa = 10.35AAYY219 pKa = 8.32PGDD222 pKa = 3.96STLSSLLRR230 pKa = 11.84AQRR233 pKa = 11.84VLKK236 pKa = 10.72GRR238 pKa = 11.84LPCGAKK244 pKa = 10.5FSRR247 pKa = 11.84NGGRR251 pKa = 11.84ASGDD255 pKa = 3.44FNTGMGNSLIMFSCVYY271 pKa = 10.72ASLVEE276 pKa = 4.34LGYY279 pKa = 10.81SSWDD283 pKa = 3.6FLCDD287 pKa = 3.36GDD289 pKa = 4.0NALLFVEE296 pKa = 5.01SSDD299 pKa = 3.79VDD301 pKa = 3.9DD302 pKa = 4.63LVEE305 pKa = 4.34RR306 pKa = 11.84LPGVAVGTCGQEE318 pKa = 3.83LSLDD322 pKa = 3.65AVTGSLSGVVFGQSKK337 pKa = 9.95PVFFPSGWRR346 pKa = 11.84LVRR349 pKa = 11.84DD350 pKa = 3.54PFKK353 pKa = 10.89ALSGFGCSHH362 pKa = 6.39AWLRR366 pKa = 11.84EE367 pKa = 3.64PKK369 pKa = 10.31FRR371 pKa = 11.84DD372 pKa = 3.46EE373 pKa = 4.3YY374 pKa = 10.51LVGVARR380 pKa = 11.84CEE382 pKa = 3.84LALSLGQPLLQAFCLRR398 pKa = 11.84VLALFPGVVPRR409 pKa = 11.84DD410 pKa = 3.34HH411 pKa = 6.97VAFRR415 pKa = 11.84DD416 pKa = 3.86YY417 pKa = 11.31EE418 pKa = 4.22FLGVSPRR425 pKa = 11.84VLRR428 pKa = 11.84RR429 pKa = 11.84PEE431 pKa = 3.82PVTWEE436 pKa = 4.19TRR438 pKa = 11.84VSFADD443 pKa = 3.46AFSISPEE450 pKa = 3.82RR451 pKa = 11.84QLEE454 pKa = 4.22IEE456 pKa = 4.24SSFDD460 pKa = 3.38LVRR463 pKa = 11.84PSGFLASDD471 pKa = 3.38AVDD474 pKa = 3.61SFFGADD480 pKa = 2.91PGMAEE485 pKa = 3.68KK486 pKa = 10.21WFEE489 pKa = 3.81RR490 pKa = 11.84WVV492 pKa = 3.31
MM1 pKa = 7.35FVPFPLPCEE10 pKa = 4.47GTWLPQVHH18 pKa = 5.87RR19 pKa = 11.84VCPHH23 pKa = 6.02NEE25 pKa = 3.31VGALCRR31 pKa = 11.84RR32 pKa = 11.84VLAPLPAQVQEE43 pKa = 4.24RR44 pKa = 11.84VSPGFSRR51 pKa = 11.84AMGLAVGLARR61 pKa = 11.84FYY63 pKa = 9.24TQPRR67 pKa = 11.84WSLEE71 pKa = 3.97RR72 pKa = 11.84TALSYY77 pKa = 11.27DD78 pKa = 3.1GALRR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 9.66LGAMDD90 pKa = 5.96SLLDD94 pKa = 3.51WPLDD98 pKa = 3.52VGKK101 pKa = 10.17DD102 pKa = 3.4SRR104 pKa = 11.84LDD106 pKa = 3.53CFLKK110 pKa = 10.84AEE112 pKa = 4.52KK113 pKa = 10.76LNTLIKK119 pKa = 10.16RR120 pKa = 11.84PKK122 pKa = 9.0PRR124 pKa = 11.84LIFPRR129 pKa = 11.84SPRR132 pKa = 11.84YY133 pKa = 9.95NLVLASRR140 pKa = 11.84LKK142 pKa = 10.71PFEE145 pKa = 3.91HH146 pKa = 6.89WLWSHH151 pKa = 6.11LTAEE155 pKa = 4.54RR156 pKa = 11.84LGTSGRR162 pKa = 11.84GRR164 pKa = 11.84YY165 pKa = 7.4VAKK168 pKa = 10.39GLNLSQRR175 pKa = 11.84ARR177 pKa = 11.84LIVKK181 pKa = 9.97KK182 pKa = 9.47MAGFSRR188 pKa = 11.84CAVLEE193 pKa = 3.99VDD195 pKa = 4.33GKK197 pKa = 11.02AFEE200 pKa = 4.27AHH202 pKa = 6.07VGPSSIEE209 pKa = 3.81QEE211 pKa = 3.88HH212 pKa = 5.84RR213 pKa = 11.84VYY215 pKa = 10.66KK216 pKa = 10.35AAYY219 pKa = 8.32PGDD222 pKa = 3.96STLSSLLRR230 pKa = 11.84AQRR233 pKa = 11.84VLKK236 pKa = 10.72GRR238 pKa = 11.84LPCGAKK244 pKa = 10.5FSRR247 pKa = 11.84NGGRR251 pKa = 11.84ASGDD255 pKa = 3.44FNTGMGNSLIMFSCVYY271 pKa = 10.72ASLVEE276 pKa = 4.34LGYY279 pKa = 10.81SSWDD283 pKa = 3.6FLCDD287 pKa = 3.36GDD289 pKa = 4.0NALLFVEE296 pKa = 5.01SSDD299 pKa = 3.79VDD301 pKa = 3.9DD302 pKa = 4.63LVEE305 pKa = 4.34RR306 pKa = 11.84LPGVAVGTCGQEE318 pKa = 3.83LSLDD322 pKa = 3.65AVTGSLSGVVFGQSKK337 pKa = 9.95PVFFPSGWRR346 pKa = 11.84LVRR349 pKa = 11.84DD350 pKa = 3.54PFKK353 pKa = 10.89ALSGFGCSHH362 pKa = 6.39AWLRR366 pKa = 11.84EE367 pKa = 3.64PKK369 pKa = 10.31FRR371 pKa = 11.84DD372 pKa = 3.46EE373 pKa = 4.3YY374 pKa = 10.51LVGVARR380 pKa = 11.84CEE382 pKa = 3.84LALSLGQPLLQAFCLRR398 pKa = 11.84VLALFPGVVPRR409 pKa = 11.84DD410 pKa = 3.34HH411 pKa = 6.97VAFRR415 pKa = 11.84DD416 pKa = 3.86YY417 pKa = 11.31EE418 pKa = 4.22FLGVSPRR425 pKa = 11.84VLRR428 pKa = 11.84RR429 pKa = 11.84PEE431 pKa = 3.82PVTWEE436 pKa = 4.19TRR438 pKa = 11.84VSFADD443 pKa = 3.46AFSISPEE450 pKa = 3.82RR451 pKa = 11.84QLEE454 pKa = 4.22IEE456 pKa = 4.24SSFDD460 pKa = 3.38LVRR463 pKa = 11.84PSGFLASDD471 pKa = 3.38AVDD474 pKa = 3.61SFFGADD480 pKa = 2.91PGMAEE485 pKa = 3.68KK486 pKa = 10.21WFEE489 pKa = 3.81RR490 pKa = 11.84WVV492 pKa = 3.31
Molecular weight: 54.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
995 |
492 |
503 |
497.5 |
55.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.04 ± 0.063 | 2.111 ± 0.229 |
4.422 ± 0.461 | 4.623 ± 0.462 |
6.935 ± 0.443 | 8.643 ± 0.216 |
1.106 ± 0.364 | 2.01 ± 0.41 |
2.714 ± 0.518 | 14.07 ± 1.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.206 ± 0.151 | 1.106 ± 0.364 |
5.628 ± 0.612 | 2.111 ± 0.229 |
8.04 ± 0.773 | 8.643 ± 0.068 |
2.714 ± 0.192 | 10.05 ± 1.058 |
3.116 ± 0.615 | 2.714 ± 0.334 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
