
Aeromicrobium marinum DSM 15272
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; Aeromicrobium marinum
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
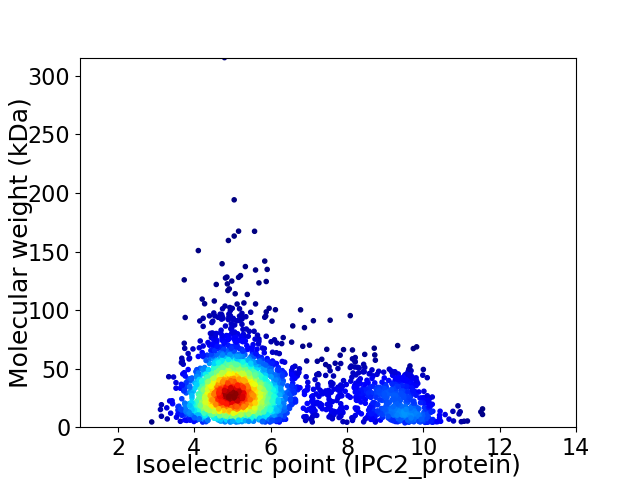
Virtual 2D-PAGE plot for 3077 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2S8S1|E2S8S1_9ACTN Uncharacterized protein OS=Aeromicrobium marinum DSM 15272 OX=585531 GN=HMPREF0063_10428 PE=4 SV=1
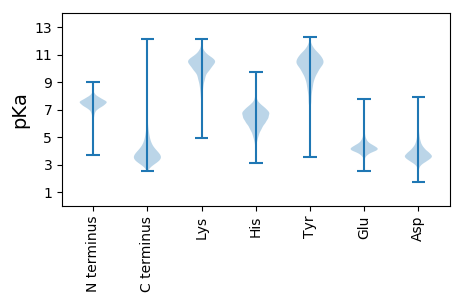
MM1 pKa = 7.55SAPDD5 pKa = 4.03PGVLDD10 pKa = 3.74RR11 pKa = 11.84RR12 pKa = 11.84ATVHH16 pKa = 6.24PPKK19 pKa = 10.45GTGTMNRR26 pKa = 11.84SLLRR30 pKa = 11.84LAVGAAAASLVLTACRR46 pKa = 11.84GDD48 pKa = 4.04DD49 pKa = 3.99GGSSAAGPGITDD61 pKa = 3.83EE62 pKa = 4.67PCPGGNADD70 pKa = 4.22NGCIYY75 pKa = 10.68LGAISDD81 pKa = 3.9LTRR84 pKa = 11.84GPFAPLAVPITDD96 pKa = 3.33AQKK99 pKa = 11.26AFWEE103 pKa = 4.36RR104 pKa = 11.84VNADD108 pKa = 3.76GGVGGYY114 pKa = 10.18DD115 pKa = 3.0INLEE119 pKa = 4.08EE120 pKa = 4.68YY121 pKa = 10.3IADD124 pKa = 3.53AEE126 pKa = 4.49YY127 pKa = 11.11NPEE130 pKa = 3.81LHH132 pKa = 6.03NRR134 pKa = 11.84EE135 pKa = 4.09YY136 pKa = 11.12QEE138 pKa = 3.75MRR140 pKa = 11.84NDD142 pKa = 3.32ILALAQTLGSSQTLAIIEE160 pKa = 4.38DD161 pKa = 3.75MRR163 pKa = 11.84SDD165 pKa = 3.84DD166 pKa = 3.88VLGVPASWNSAWNFDD181 pKa = 4.01DD182 pKa = 5.57QILEE186 pKa = 4.19SGANYY191 pKa = 10.05CFEE194 pKa = 4.52AMNGVDD200 pKa = 2.9WAVAEE205 pKa = 4.33RR206 pKa = 11.84GVSEE210 pKa = 4.09TVLAVGYY217 pKa = 9.35PGDD220 pKa = 3.87YY221 pKa = 11.03GGDD224 pKa = 3.12AAAGVEE230 pKa = 4.25AAAEE234 pKa = 4.09ANGLEE239 pKa = 4.4FSQVEE244 pKa = 4.4TPPGQDD250 pKa = 2.99NQAGAIQQILAQQPDD265 pKa = 4.05LVFISTGPAEE275 pKa = 4.18MATIVGGSAAQGFTGTFVGSSPTWNPALLQSAAAPALEE313 pKa = 4.09ALYY316 pKa = 9.95FQAGPWGPFGTEE328 pKa = 3.93TDD330 pKa = 2.87GHH332 pKa = 6.35AAMRR336 pKa = 11.84DD337 pKa = 3.37ALGDD341 pKa = 3.48VTPNDD346 pKa = 4.48GYY348 pKa = 8.88TAGWAWSYY356 pKa = 11.3PLLAALEE363 pKa = 4.3AAAEE367 pKa = 4.02LDD369 pKa = 3.76GGITRR374 pKa = 11.84EE375 pKa = 3.99NLLAAAADD383 pKa = 3.97LEE385 pKa = 4.41EE386 pKa = 4.36VDD388 pKa = 4.39YY389 pKa = 11.7EE390 pKa = 4.64GMLPNEE396 pKa = 4.6AGNYY400 pKa = 9.94AGDD403 pKa = 3.77PSEE406 pKa = 4.01VAFRR410 pKa = 11.84QSVISGVDD418 pKa = 3.15TSAPTGVAIVQEE430 pKa = 4.7FQAGPTATDD439 pKa = 3.49YY440 pKa = 11.53DD441 pKa = 4.36FSSPCFSLNN450 pKa = 3.1
MM1 pKa = 7.55SAPDD5 pKa = 4.03PGVLDD10 pKa = 3.74RR11 pKa = 11.84RR12 pKa = 11.84ATVHH16 pKa = 6.24PPKK19 pKa = 10.45GTGTMNRR26 pKa = 11.84SLLRR30 pKa = 11.84LAVGAAAASLVLTACRR46 pKa = 11.84GDD48 pKa = 4.04DD49 pKa = 3.99GGSSAAGPGITDD61 pKa = 3.83EE62 pKa = 4.67PCPGGNADD70 pKa = 4.22NGCIYY75 pKa = 10.68LGAISDD81 pKa = 3.9LTRR84 pKa = 11.84GPFAPLAVPITDD96 pKa = 3.33AQKK99 pKa = 11.26AFWEE103 pKa = 4.36RR104 pKa = 11.84VNADD108 pKa = 3.76GGVGGYY114 pKa = 10.18DD115 pKa = 3.0INLEE119 pKa = 4.08EE120 pKa = 4.68YY121 pKa = 10.3IADD124 pKa = 3.53AEE126 pKa = 4.49YY127 pKa = 11.11NPEE130 pKa = 3.81LHH132 pKa = 6.03NRR134 pKa = 11.84EE135 pKa = 4.09YY136 pKa = 11.12QEE138 pKa = 3.75MRR140 pKa = 11.84NDD142 pKa = 3.32ILALAQTLGSSQTLAIIEE160 pKa = 4.38DD161 pKa = 3.75MRR163 pKa = 11.84SDD165 pKa = 3.84DD166 pKa = 3.88VLGVPASWNSAWNFDD181 pKa = 4.01DD182 pKa = 5.57QILEE186 pKa = 4.19SGANYY191 pKa = 10.05CFEE194 pKa = 4.52AMNGVDD200 pKa = 2.9WAVAEE205 pKa = 4.33RR206 pKa = 11.84GVSEE210 pKa = 4.09TVLAVGYY217 pKa = 9.35PGDD220 pKa = 3.87YY221 pKa = 11.03GGDD224 pKa = 3.12AAAGVEE230 pKa = 4.25AAAEE234 pKa = 4.09ANGLEE239 pKa = 4.4FSQVEE244 pKa = 4.4TPPGQDD250 pKa = 2.99NQAGAIQQILAQQPDD265 pKa = 4.05LVFISTGPAEE275 pKa = 4.18MATIVGGSAAQGFTGTFVGSSPTWNPALLQSAAAPALEE313 pKa = 4.09ALYY316 pKa = 9.95FQAGPWGPFGTEE328 pKa = 3.93TDD330 pKa = 2.87GHH332 pKa = 6.35AAMRR336 pKa = 11.84DD337 pKa = 3.37ALGDD341 pKa = 3.48VTPNDD346 pKa = 4.48GYY348 pKa = 8.88TAGWAWSYY356 pKa = 11.3PLLAALEE363 pKa = 4.3AAAEE367 pKa = 4.02LDD369 pKa = 3.76GGITRR374 pKa = 11.84EE375 pKa = 3.99NLLAAAADD383 pKa = 3.97LEE385 pKa = 4.41EE386 pKa = 4.36VDD388 pKa = 4.39YY389 pKa = 11.7EE390 pKa = 4.64GMLPNEE396 pKa = 4.6AGNYY400 pKa = 9.94AGDD403 pKa = 3.77PSEE406 pKa = 4.01VAFRR410 pKa = 11.84QSVISGVDD418 pKa = 3.15TSAPTGVAIVQEE430 pKa = 4.7FQAGPTATDD439 pKa = 3.49YY440 pKa = 11.53DD441 pKa = 4.36FSSPCFSLNN450 pKa = 3.1
Molecular weight: 46.57 kDa
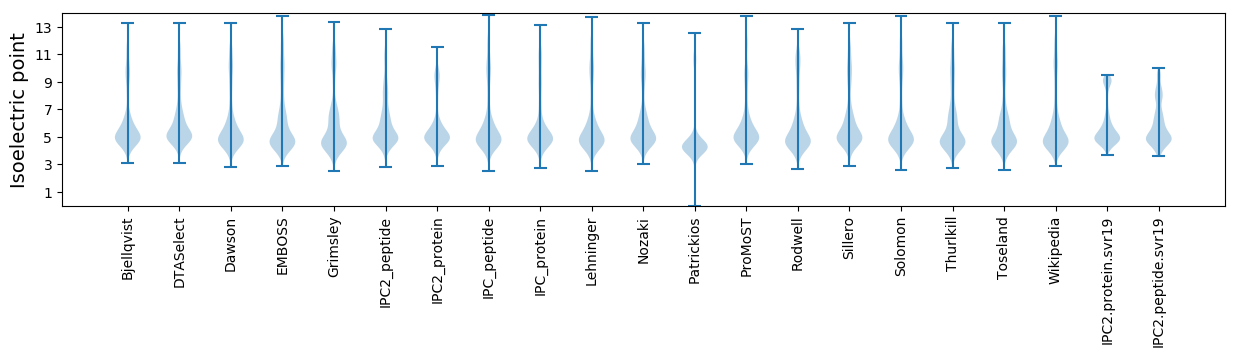
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2S974|E2S974_9ACTN ENDO3c domain-containing protein OS=Aeromicrobium marinum DSM 15272 OX=585531 GN=HMPREF0063_10514 PE=4 SV=1
MM1 pKa = 7.24ATLPRR6 pKa = 11.84TTRR9 pKa = 11.84SSPVATPLRR18 pKa = 11.84TTPRR22 pKa = 11.84SPVATLPRR30 pKa = 11.84TTPSLPVATLPRR42 pKa = 11.84TAPRR46 pKa = 11.84SATPTTPTTPTTRR59 pKa = 11.84TTRR62 pKa = 11.84TTPPRR67 pKa = 11.84TTTVSTSRR75 pKa = 11.84RR76 pKa = 11.84ATTTPVRR83 pKa = 11.84TTPPRR88 pKa = 11.84TTMVSTSRR96 pKa = 11.84RR97 pKa = 11.84ATTTPVRR104 pKa = 11.84TTPTTPTTRR113 pKa = 11.84TTRR116 pKa = 11.84TTPTTRR122 pKa = 3.16
MM1 pKa = 7.24ATLPRR6 pKa = 11.84TTRR9 pKa = 11.84SSPVATPLRR18 pKa = 11.84TTPRR22 pKa = 11.84SPVATLPRR30 pKa = 11.84TTPSLPVATLPRR42 pKa = 11.84TAPRR46 pKa = 11.84SATPTTPTTPTTRR59 pKa = 11.84TTRR62 pKa = 11.84TTPPRR67 pKa = 11.84TTTVSTSRR75 pKa = 11.84RR76 pKa = 11.84ATTTPVRR83 pKa = 11.84TTPPRR88 pKa = 11.84TTMVSTSRR96 pKa = 11.84RR97 pKa = 11.84ATTTPVRR104 pKa = 11.84TTPTTPTTRR113 pKa = 11.84TTRR116 pKa = 11.84TTPTTRR122 pKa = 3.16
Molecular weight: 13.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
937159 |
31 |
3019 |
304.6 |
32.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.934 ± 0.055 | 0.693 ± 0.011 |
6.778 ± 0.035 | 5.705 ± 0.041 |
2.932 ± 0.023 | 9.18 ± 0.039 |
2.214 ± 0.022 | 3.717 ± 0.033 |
1.713 ± 0.032 | 9.97 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.857 ± 0.018 | 1.692 ± 0.023 |
5.56 ± 0.038 | 2.748 ± 0.022 |
7.773 ± 0.049 | 5.277 ± 0.027 |
6.286 ± 0.035 | 9.696 ± 0.05 |
1.469 ± 0.02 | 1.808 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
