
Prevotella sp. CAG:1320
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Prevotellaceae; Prevotella; environmental samples
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
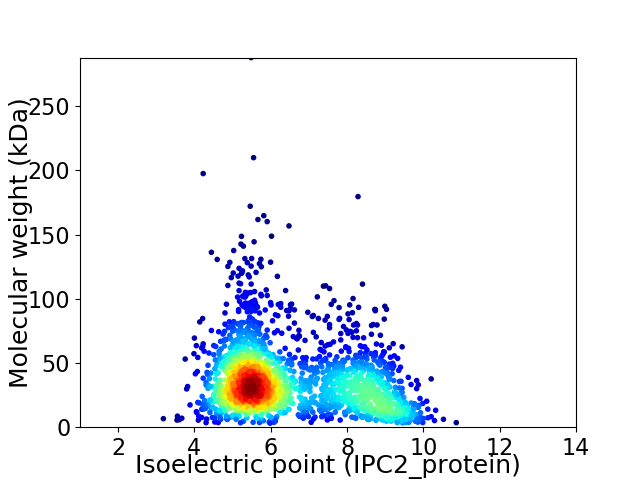
Virtual 2D-PAGE plot for 2035 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6EVT0|R6EVT0_9BACT Uncharacterized protein OS=Prevotella sp. CAG:1320 OX=1262922 GN=BN487_00768 PE=4 SV=1
MM1 pKa = 7.86KK2 pKa = 10.12DD3 pKa = 3.44LKK5 pKa = 10.81VLKK8 pKa = 10.5LSIPTLDD15 pKa = 3.37SSEE18 pKa = 4.73CKK20 pKa = 9.89TLMGGDD26 pKa = 3.7GYY28 pKa = 11.27GYY30 pKa = 10.72DD31 pKa = 4.14VIGGDD36 pKa = 3.77LQEE39 pKa = 4.73CVITADD45 pKa = 4.58GYY47 pKa = 11.31RR48 pKa = 11.84PDD50 pKa = 4.03HH51 pKa = 7.1SDD53 pKa = 3.39YY54 pKa = 11.13DD55 pKa = 4.21YY56 pKa = 11.77QDD58 pKa = 3.54DD59 pKa = 4.61TLDD62 pKa = 5.34DD63 pKa = 3.88EE64 pKa = 4.74QNDD67 pKa = 3.5YY68 pKa = 11.39GYY70 pKa = 11.29DD71 pKa = 3.41HH72 pKa = 7.85DD73 pKa = 5.51YY74 pKa = 11.79DD75 pKa = 3.82NSHH78 pKa = 7.34DD79 pKa = 3.85NDD81 pKa = 4.05YY82 pKa = 11.46NVPDD86 pKa = 3.61QLKK89 pKa = 8.23GTFEE93 pKa = 4.25KK94 pKa = 10.82LPEE97 pKa = 4.79KK98 pKa = 10.6IQDD101 pKa = 3.5FLKK104 pKa = 10.98ANNIKK109 pKa = 10.19IIYY112 pKa = 9.09DD113 pKa = 3.52PNYY116 pKa = 9.88KK117 pKa = 10.24NGSGGPASYY126 pKa = 10.33FSEE129 pKa = 4.42DD130 pKa = 3.28KK131 pKa = 10.96SIKK134 pKa = 9.67TSSLDD139 pKa = 3.56DD140 pKa = 4.6NILLRR145 pKa = 11.84EE146 pKa = 4.26VIHH149 pKa = 6.62AVQDD153 pKa = 3.55AMGSLDD159 pKa = 3.87GMSHH163 pKa = 5.85SAEE166 pKa = 4.05EE167 pKa = 4.26FQEE170 pKa = 4.29KK171 pKa = 10.68ALGDD175 pKa = 3.57LASYY179 pKa = 10.45FNGVINEE186 pKa = 4.08NGGCGFTLDD195 pKa = 4.36FSDD198 pKa = 5.67GNSSWSEE205 pKa = 3.71FVGSCFDD212 pKa = 5.36DD213 pKa = 4.89DD214 pKa = 6.07DD215 pKa = 5.16NFDD218 pKa = 3.51RR219 pKa = 11.84DD220 pKa = 3.64AFKK223 pKa = 10.95EE224 pKa = 4.5GIMDD228 pKa = 4.55FFDD231 pKa = 3.87IFQDD235 pKa = 3.28THH237 pKa = 8.23SNVSGYY243 pKa = 10.76NDD245 pKa = 3.52SLEE248 pKa = 5.64DD249 pKa = 3.45NYY251 pKa = 11.26NWHH254 pKa = 6.54WDD256 pKa = 3.54EE257 pKa = 5.62FFDD260 pKa = 5.82LLGLL264 pKa = 4.05
MM1 pKa = 7.86KK2 pKa = 10.12DD3 pKa = 3.44LKK5 pKa = 10.81VLKK8 pKa = 10.5LSIPTLDD15 pKa = 3.37SSEE18 pKa = 4.73CKK20 pKa = 9.89TLMGGDD26 pKa = 3.7GYY28 pKa = 11.27GYY30 pKa = 10.72DD31 pKa = 4.14VIGGDD36 pKa = 3.77LQEE39 pKa = 4.73CVITADD45 pKa = 4.58GYY47 pKa = 11.31RR48 pKa = 11.84PDD50 pKa = 4.03HH51 pKa = 7.1SDD53 pKa = 3.39YY54 pKa = 11.13DD55 pKa = 4.21YY56 pKa = 11.77QDD58 pKa = 3.54DD59 pKa = 4.61TLDD62 pKa = 5.34DD63 pKa = 3.88EE64 pKa = 4.74QNDD67 pKa = 3.5YY68 pKa = 11.39GYY70 pKa = 11.29DD71 pKa = 3.41HH72 pKa = 7.85DD73 pKa = 5.51YY74 pKa = 11.79DD75 pKa = 3.82NSHH78 pKa = 7.34DD79 pKa = 3.85NDD81 pKa = 4.05YY82 pKa = 11.46NVPDD86 pKa = 3.61QLKK89 pKa = 8.23GTFEE93 pKa = 4.25KK94 pKa = 10.82LPEE97 pKa = 4.79KK98 pKa = 10.6IQDD101 pKa = 3.5FLKK104 pKa = 10.98ANNIKK109 pKa = 10.19IIYY112 pKa = 9.09DD113 pKa = 3.52PNYY116 pKa = 9.88KK117 pKa = 10.24NGSGGPASYY126 pKa = 10.33FSEE129 pKa = 4.42DD130 pKa = 3.28KK131 pKa = 10.96SIKK134 pKa = 9.67TSSLDD139 pKa = 3.56DD140 pKa = 4.6NILLRR145 pKa = 11.84EE146 pKa = 4.26VIHH149 pKa = 6.62AVQDD153 pKa = 3.55AMGSLDD159 pKa = 3.87GMSHH163 pKa = 5.85SAEE166 pKa = 4.05EE167 pKa = 4.26FQEE170 pKa = 4.29KK171 pKa = 10.68ALGDD175 pKa = 3.57LASYY179 pKa = 10.45FNGVINEE186 pKa = 4.08NGGCGFTLDD195 pKa = 4.36FSDD198 pKa = 5.67GNSSWSEE205 pKa = 3.71FVGSCFDD212 pKa = 5.36DD213 pKa = 4.89DD214 pKa = 6.07DD215 pKa = 5.16NFDD218 pKa = 3.51RR219 pKa = 11.84DD220 pKa = 3.64AFKK223 pKa = 10.95EE224 pKa = 4.5GIMDD228 pKa = 4.55FFDD231 pKa = 3.87IFQDD235 pKa = 3.28THH237 pKa = 8.23SNVSGYY243 pKa = 10.76NDD245 pKa = 3.52SLEE248 pKa = 5.64DD249 pKa = 3.45NYY251 pKa = 11.26NWHH254 pKa = 6.54WDD256 pKa = 3.54EE257 pKa = 5.62FFDD260 pKa = 5.82LLGLL264 pKa = 4.05
Molecular weight: 29.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6E1Y2|R6E1Y2_9BACT Coenzyme A biosynthesis bifunctional protein CoaBC OS=Prevotella sp. CAG:1320 OX=1262922 GN=coaBC PE=3 SV=1
MM1 pKa = 7.85PNGKK5 pKa = 9.19KK6 pKa = 10.25KK7 pKa = 10.12KK8 pKa = 7.0GHH10 pKa = 6.14KK11 pKa = 9.06MATHH15 pKa = 6.13KK16 pKa = 10.39RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.25NRR25 pKa = 11.84HH26 pKa = 4.69KK27 pKa = 11.1SKK29 pKa = 11.1
MM1 pKa = 7.85PNGKK5 pKa = 9.19KK6 pKa = 10.25KK7 pKa = 10.12KK8 pKa = 7.0GHH10 pKa = 6.14KK11 pKa = 9.06MATHH15 pKa = 6.13KK16 pKa = 10.39RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.25NRR25 pKa = 11.84HH26 pKa = 4.69KK27 pKa = 11.1SKK29 pKa = 11.1
Molecular weight: 3.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
691449 |
29 |
2543 |
339.8 |
38.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.807 ± 0.049 | 1.329 ± 0.017 |
5.755 ± 0.039 | 6.578 ± 0.051 |
4.206 ± 0.034 | 7.088 ± 0.045 |
2.144 ± 0.023 | 6.122 ± 0.046 |
6.027 ± 0.049 | 9.168 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.993 ± 0.025 | 4.424 ± 0.048 |
3.866 ± 0.028 | 3.626 ± 0.027 |
5.38 ± 0.044 | 5.648 ± 0.043 |
5.47 ± 0.04 | 6.829 ± 0.044 |
1.282 ± 0.024 | 4.254 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
