
Penicillium stoloniferum virus S
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Gammapartitivirus
Average proteome isoelectric point is 7.42
Get precalculated fractions of proteins
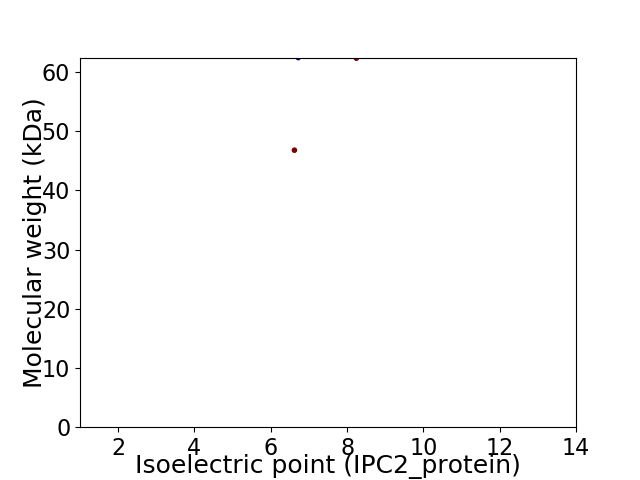
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6YDQ7|Q6YDQ7_9VIRU RNA-dependent RNA polymerase OS=Penicillium stoloniferum virus S OX=216371 PE=4 SV=2
MM1 pKa = 7.51SSIAPTDD8 pKa = 3.89SVSSSGKK15 pKa = 8.95RR16 pKa = 11.84SKK18 pKa = 10.08PGKK21 pKa = 9.7RR22 pKa = 11.84EE23 pKa = 3.7RR24 pKa = 11.84QQARR28 pKa = 11.84SAVGSAGGKK37 pKa = 8.09PASASKK43 pKa = 10.42AAAFAQGGSSDD54 pKa = 4.2PVPMPGKK61 pKa = 9.55YY62 pKa = 9.27PVVFSTGAGEE72 pKa = 3.96PTRR75 pKa = 11.84DD76 pKa = 3.34QEE78 pKa = 4.59FALPVHH84 pKa = 6.38KK85 pKa = 10.51AFPLFGSVSDD95 pKa = 3.79KK96 pKa = 10.51YY97 pKa = 10.86RR98 pKa = 11.84RR99 pKa = 11.84NPRR102 pKa = 11.84YY103 pKa = 10.13AEE105 pKa = 4.01FRR107 pKa = 11.84AHH109 pKa = 7.24SEE111 pKa = 3.85FTDD114 pKa = 4.5GVFGTHH120 pKa = 6.65LAVSSLLRR128 pKa = 11.84LAQQLVHH135 pKa = 6.26AHH137 pKa = 5.26VNMGLPLGDD146 pKa = 4.44FAPLASSDD154 pKa = 3.46VRR156 pKa = 11.84IPSALASVVNQFGEE170 pKa = 4.48FSSPSIGTRR179 pKa = 11.84FLLRR183 pKa = 11.84DD184 pKa = 3.45YY185 pKa = 9.6EE186 pKa = 4.44HH187 pKa = 6.79AVSRR191 pKa = 11.84VVFLADD197 pKa = 3.8QLWTNGNSHH206 pKa = 7.21HH207 pKa = 6.5IFARR211 pKa = 11.84SWLPMSNNDD220 pKa = 3.6GNFKK224 pKa = 10.39TIVASRR230 pKa = 11.84LLEE233 pKa = 4.65FISAGDD239 pKa = 3.68LSILPTVLEE248 pKa = 4.33DD249 pKa = 3.41AVLSGEE255 pKa = 4.23VPEE258 pKa = 4.98AWEE261 pKa = 3.96QVKK264 pKa = 10.74DD265 pKa = 4.08LLGDD269 pKa = 3.7APGVGQVDD277 pKa = 3.11RR278 pKa = 11.84RR279 pKa = 11.84DD280 pKa = 3.44RR281 pKa = 11.84FDD283 pKa = 5.13FLFKK287 pKa = 10.7SYY289 pKa = 11.52ADD291 pKa = 3.29VGQFTTAFTTQAASDD306 pKa = 3.84VLTEE310 pKa = 6.13LGLPWNSPSAGHH322 pKa = 7.03LNWQYY327 pKa = 9.3STKK330 pKa = 10.3QRR332 pKa = 11.84FTFLADD338 pKa = 3.24TWAKK342 pKa = 10.61LSAAYY347 pKa = 10.16SQFFEE352 pKa = 4.74LSSGLATRR360 pKa = 11.84QSATGSHH367 pKa = 5.79AQMVDD372 pKa = 3.26LTSVEE377 pKa = 4.19GVTVLKK383 pKa = 10.71AALALSAPEE392 pKa = 4.39FSLAACFPPSCIFVGGITRR411 pKa = 11.84RR412 pKa = 11.84VVVTTSLSVSQRR424 pKa = 11.84ATEE427 pKa = 4.57FCQMDD432 pKa = 3.2WRR434 pKa = 5.42
MM1 pKa = 7.51SSIAPTDD8 pKa = 3.89SVSSSGKK15 pKa = 8.95RR16 pKa = 11.84SKK18 pKa = 10.08PGKK21 pKa = 9.7RR22 pKa = 11.84EE23 pKa = 3.7RR24 pKa = 11.84QQARR28 pKa = 11.84SAVGSAGGKK37 pKa = 8.09PASASKK43 pKa = 10.42AAAFAQGGSSDD54 pKa = 4.2PVPMPGKK61 pKa = 9.55YY62 pKa = 9.27PVVFSTGAGEE72 pKa = 3.96PTRR75 pKa = 11.84DD76 pKa = 3.34QEE78 pKa = 4.59FALPVHH84 pKa = 6.38KK85 pKa = 10.51AFPLFGSVSDD95 pKa = 3.79KK96 pKa = 10.51YY97 pKa = 10.86RR98 pKa = 11.84RR99 pKa = 11.84NPRR102 pKa = 11.84YY103 pKa = 10.13AEE105 pKa = 4.01FRR107 pKa = 11.84AHH109 pKa = 7.24SEE111 pKa = 3.85FTDD114 pKa = 4.5GVFGTHH120 pKa = 6.65LAVSSLLRR128 pKa = 11.84LAQQLVHH135 pKa = 6.26AHH137 pKa = 5.26VNMGLPLGDD146 pKa = 4.44FAPLASSDD154 pKa = 3.46VRR156 pKa = 11.84IPSALASVVNQFGEE170 pKa = 4.48FSSPSIGTRR179 pKa = 11.84FLLRR183 pKa = 11.84DD184 pKa = 3.45YY185 pKa = 9.6EE186 pKa = 4.44HH187 pKa = 6.79AVSRR191 pKa = 11.84VVFLADD197 pKa = 3.8QLWTNGNSHH206 pKa = 7.21HH207 pKa = 6.5IFARR211 pKa = 11.84SWLPMSNNDD220 pKa = 3.6GNFKK224 pKa = 10.39TIVASRR230 pKa = 11.84LLEE233 pKa = 4.65FISAGDD239 pKa = 3.68LSILPTVLEE248 pKa = 4.33DD249 pKa = 3.41AVLSGEE255 pKa = 4.23VPEE258 pKa = 4.98AWEE261 pKa = 3.96QVKK264 pKa = 10.74DD265 pKa = 4.08LLGDD269 pKa = 3.7APGVGQVDD277 pKa = 3.11RR278 pKa = 11.84RR279 pKa = 11.84DD280 pKa = 3.44RR281 pKa = 11.84FDD283 pKa = 5.13FLFKK287 pKa = 10.7SYY289 pKa = 11.52ADD291 pKa = 3.29VGQFTTAFTTQAASDD306 pKa = 3.84VLTEE310 pKa = 6.13LGLPWNSPSAGHH322 pKa = 7.03LNWQYY327 pKa = 9.3STKK330 pKa = 10.3QRR332 pKa = 11.84FTFLADD338 pKa = 3.24TWAKK342 pKa = 10.61LSAAYY347 pKa = 10.16SQFFEE352 pKa = 4.74LSSGLATRR360 pKa = 11.84QSATGSHH367 pKa = 5.79AQMVDD372 pKa = 3.26LTSVEE377 pKa = 4.19GVTVLKK383 pKa = 10.71AALALSAPEE392 pKa = 4.39FSLAACFPPSCIFVGGITRR411 pKa = 11.84RR412 pKa = 11.84VVVTTSLSVSQRR424 pKa = 11.84ATEE427 pKa = 4.57FCQMDD432 pKa = 3.2WRR434 pKa = 5.42
Molecular weight: 46.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6YDQ7|Q6YDQ7_9VIRU RNA-dependent RNA polymerase OS=Penicillium stoloniferum virus S OX=216371 PE=4 SV=2
MM1 pKa = 7.81EE2 pKa = 6.64DD3 pKa = 3.67SPFDD7 pKa = 3.66PTLLDD12 pKa = 3.48VAVEE16 pKa = 4.08EE17 pKa = 4.37SHH19 pKa = 7.24LVDD22 pKa = 5.76DD23 pKa = 5.71SPLTPSNRR31 pKa = 11.84TRR33 pKa = 11.84GASYY37 pKa = 10.61GIISEE42 pKa = 4.51KK43 pKa = 10.35FSSPGLRR50 pKa = 11.84EE51 pKa = 3.35IARR54 pKa = 11.84YY55 pKa = 9.42GGYY58 pKa = 9.58SVYY61 pKa = 10.57SGQSNTDD68 pKa = 2.27AWIRR72 pKa = 11.84TTLKK76 pKa = 10.12EE77 pKa = 3.99FDD79 pKa = 3.29RR80 pKa = 11.84SVYY83 pKa = 10.91DD84 pKa = 4.61DD85 pKa = 3.2IYY87 pKa = 10.86GYY89 pKa = 7.49TRR91 pKa = 11.84RR92 pKa = 11.84PTGTLGMYY100 pKa = 10.6GSLLKK105 pKa = 10.71FSEE108 pKa = 4.48GKK110 pKa = 10.39NSFASLNRR118 pKa = 11.84VQRR121 pKa = 11.84KK122 pKa = 8.99SMINAISKK130 pKa = 9.92AKK132 pKa = 10.2KK133 pKa = 9.99AFKK136 pKa = 10.37LPYY139 pKa = 9.5QRR141 pKa = 11.84EE142 pKa = 3.99PLDD145 pKa = 3.08WHH147 pKa = 5.46EE148 pKa = 4.44VGRR151 pKa = 11.84HH152 pKa = 4.2FRR154 pKa = 11.84RR155 pKa = 11.84DD156 pKa = 3.05TSAGVSFMGQKK167 pKa = 10.24KK168 pKa = 9.61GDD170 pKa = 3.65VMEE173 pKa = 5.22EE174 pKa = 4.04IYY176 pKa = 10.96HH177 pKa = 5.05EE178 pKa = 4.4ARR180 pKa = 11.84WLGHH184 pKa = 5.84RR185 pKa = 11.84MKK187 pKa = 10.97QNGRR191 pKa = 11.84AKK193 pKa = 10.37FDD195 pKa = 3.77PSKK198 pKa = 10.71MRR200 pKa = 11.84FPPCLAGQRR209 pKa = 11.84GGMSEE214 pKa = 4.39ASDD217 pKa = 3.44PKK219 pKa = 10.64TRR221 pKa = 11.84LVWIYY226 pKa = 9.11PAEE229 pKa = 3.94MLAIEE234 pKa = 5.05GFYY237 pKa = 10.75APEE240 pKa = 4.22MYY242 pKa = 9.71HH243 pKa = 8.87AYY245 pKa = 9.47MDD247 pKa = 4.96DD248 pKa = 3.94PLSPMLNGKK257 pKa = 9.39SSQRR261 pKa = 11.84LYY263 pKa = 10.75TEE265 pKa = 4.47WTCGLRR271 pKa = 11.84DD272 pKa = 3.36GEE274 pKa = 4.33MLYY277 pKa = 11.21GLDD280 pKa = 4.39FSGFDD285 pKa = 3.33TKK287 pKa = 11.07VPAWLIRR294 pKa = 11.84VAFDD298 pKa = 3.86ILRR301 pKa = 11.84QNIRR305 pKa = 11.84WDD307 pKa = 3.36SFRR310 pKa = 11.84GEE312 pKa = 4.2KK313 pKa = 9.8VSKK316 pKa = 10.5RR317 pKa = 11.84DD318 pKa = 3.4AQKK321 pKa = 10.19WRR323 pKa = 11.84NVWDD327 pKa = 3.58GMVWYY332 pKa = 8.64FINTPILMPDD342 pKa = 2.41GRR344 pKa = 11.84MFRR347 pKa = 11.84KK348 pKa = 9.98YY349 pKa = 10.14RR350 pKa = 11.84GVPSGSWFTQMIDD363 pKa = 3.34SVVNYY368 pKa = 10.2ILVDD372 pKa = 3.46YY373 pKa = 10.57LAACQQCEE381 pKa = 3.73IRR383 pKa = 11.84ALRR386 pKa = 11.84VLGDD390 pKa = 3.57DD391 pKa = 3.83SAFRR395 pKa = 11.84SCDD398 pKa = 3.48PFSLDD403 pKa = 4.39LASHH407 pKa = 6.92DD408 pKa = 4.04AEE410 pKa = 4.35CVNMILHH417 pKa = 6.69PEE419 pKa = 3.98KK420 pKa = 10.75CEE422 pKa = 3.91KK423 pKa = 10.35TKK425 pKa = 11.24DD426 pKa = 3.27PTAFKK431 pKa = 11.03LLGTTYY437 pKa = 10.92RR438 pKa = 11.84NGRR441 pKa = 11.84PHH443 pKa = 7.54RR444 pKa = 11.84EE445 pKa = 3.68TNEE448 pKa = 3.71WFKK451 pKa = 11.1LALYY455 pKa = 9.01PEE457 pKa = 4.62SVVPSLQVSFTRR469 pKa = 11.84LIGLWIGGAMFDD481 pKa = 4.74SRR483 pKa = 11.84FCQFMEE489 pKa = 4.47YY490 pKa = 10.51YY491 pKa = 8.36QTCFPCPQEE500 pKa = 3.82GWFSKK505 pKa = 9.82DD506 pKa = 3.0QRR508 pKa = 11.84RR509 pKa = 11.84WLQVVYY515 pKa = 10.42GGKK518 pKa = 10.08APRR521 pKa = 11.84GWTTKK526 pKa = 10.44RR527 pKa = 11.84SLFWRR532 pKa = 11.84SIFYY536 pKa = 10.87VYY538 pKa = 10.99GG539 pKa = 3.59
MM1 pKa = 7.81EE2 pKa = 6.64DD3 pKa = 3.67SPFDD7 pKa = 3.66PTLLDD12 pKa = 3.48VAVEE16 pKa = 4.08EE17 pKa = 4.37SHH19 pKa = 7.24LVDD22 pKa = 5.76DD23 pKa = 5.71SPLTPSNRR31 pKa = 11.84TRR33 pKa = 11.84GASYY37 pKa = 10.61GIISEE42 pKa = 4.51KK43 pKa = 10.35FSSPGLRR50 pKa = 11.84EE51 pKa = 3.35IARR54 pKa = 11.84YY55 pKa = 9.42GGYY58 pKa = 9.58SVYY61 pKa = 10.57SGQSNTDD68 pKa = 2.27AWIRR72 pKa = 11.84TTLKK76 pKa = 10.12EE77 pKa = 3.99FDD79 pKa = 3.29RR80 pKa = 11.84SVYY83 pKa = 10.91DD84 pKa = 4.61DD85 pKa = 3.2IYY87 pKa = 10.86GYY89 pKa = 7.49TRR91 pKa = 11.84RR92 pKa = 11.84PTGTLGMYY100 pKa = 10.6GSLLKK105 pKa = 10.71FSEE108 pKa = 4.48GKK110 pKa = 10.39NSFASLNRR118 pKa = 11.84VQRR121 pKa = 11.84KK122 pKa = 8.99SMINAISKK130 pKa = 9.92AKK132 pKa = 10.2KK133 pKa = 9.99AFKK136 pKa = 10.37LPYY139 pKa = 9.5QRR141 pKa = 11.84EE142 pKa = 3.99PLDD145 pKa = 3.08WHH147 pKa = 5.46EE148 pKa = 4.44VGRR151 pKa = 11.84HH152 pKa = 4.2FRR154 pKa = 11.84RR155 pKa = 11.84DD156 pKa = 3.05TSAGVSFMGQKK167 pKa = 10.24KK168 pKa = 9.61GDD170 pKa = 3.65VMEE173 pKa = 5.22EE174 pKa = 4.04IYY176 pKa = 10.96HH177 pKa = 5.05EE178 pKa = 4.4ARR180 pKa = 11.84WLGHH184 pKa = 5.84RR185 pKa = 11.84MKK187 pKa = 10.97QNGRR191 pKa = 11.84AKK193 pKa = 10.37FDD195 pKa = 3.77PSKK198 pKa = 10.71MRR200 pKa = 11.84FPPCLAGQRR209 pKa = 11.84GGMSEE214 pKa = 4.39ASDD217 pKa = 3.44PKK219 pKa = 10.64TRR221 pKa = 11.84LVWIYY226 pKa = 9.11PAEE229 pKa = 3.94MLAIEE234 pKa = 5.05GFYY237 pKa = 10.75APEE240 pKa = 4.22MYY242 pKa = 9.71HH243 pKa = 8.87AYY245 pKa = 9.47MDD247 pKa = 4.96DD248 pKa = 3.94PLSPMLNGKK257 pKa = 9.39SSQRR261 pKa = 11.84LYY263 pKa = 10.75TEE265 pKa = 4.47WTCGLRR271 pKa = 11.84DD272 pKa = 3.36GEE274 pKa = 4.33MLYY277 pKa = 11.21GLDD280 pKa = 4.39FSGFDD285 pKa = 3.33TKK287 pKa = 11.07VPAWLIRR294 pKa = 11.84VAFDD298 pKa = 3.86ILRR301 pKa = 11.84QNIRR305 pKa = 11.84WDD307 pKa = 3.36SFRR310 pKa = 11.84GEE312 pKa = 4.2KK313 pKa = 9.8VSKK316 pKa = 10.5RR317 pKa = 11.84DD318 pKa = 3.4AQKK321 pKa = 10.19WRR323 pKa = 11.84NVWDD327 pKa = 3.58GMVWYY332 pKa = 8.64FINTPILMPDD342 pKa = 2.41GRR344 pKa = 11.84MFRR347 pKa = 11.84KK348 pKa = 9.98YY349 pKa = 10.14RR350 pKa = 11.84GVPSGSWFTQMIDD363 pKa = 3.34SVVNYY368 pKa = 10.2ILVDD372 pKa = 3.46YY373 pKa = 10.57LAACQQCEE381 pKa = 3.73IRR383 pKa = 11.84ALRR386 pKa = 11.84VLGDD390 pKa = 3.57DD391 pKa = 3.83SAFRR395 pKa = 11.84SCDD398 pKa = 3.48PFSLDD403 pKa = 4.39LASHH407 pKa = 6.92DD408 pKa = 4.04AEE410 pKa = 4.35CVNMILHH417 pKa = 6.69PEE419 pKa = 3.98KK420 pKa = 10.75CEE422 pKa = 3.91KK423 pKa = 10.35TKK425 pKa = 11.24DD426 pKa = 3.27PTAFKK431 pKa = 11.03LLGTTYY437 pKa = 10.92RR438 pKa = 11.84NGRR441 pKa = 11.84PHH443 pKa = 7.54RR444 pKa = 11.84EE445 pKa = 3.68TNEE448 pKa = 3.71WFKK451 pKa = 11.1LALYY455 pKa = 9.01PEE457 pKa = 4.62SVVPSLQVSFTRR469 pKa = 11.84LIGLWIGGAMFDD481 pKa = 4.74SRR483 pKa = 11.84FCQFMEE489 pKa = 4.47YY490 pKa = 10.51YY491 pKa = 8.36QTCFPCPQEE500 pKa = 3.82GWFSKK505 pKa = 9.82DD506 pKa = 3.0QRR508 pKa = 11.84RR509 pKa = 11.84WLQVVYY515 pKa = 10.42GGKK518 pKa = 10.08APRR521 pKa = 11.84GWTTKK526 pKa = 10.44RR527 pKa = 11.84SLFWRR532 pKa = 11.84SIFYY536 pKa = 10.87VYY538 pKa = 10.99GG539 pKa = 3.59
Molecular weight: 62.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
973 |
434 |
539 |
486.5 |
54.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.119 ± 2.071 | 1.336 ± 0.421 |
5.755 ± 0.448 | 4.625 ± 0.462 |
6.064 ± 0.404 | 7.605 ± 0.152 |
1.953 ± 0.229 | 3.186 ± 0.726 |
4.419 ± 0.779 | 8.222 ± 0.499 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.672 ± 0.842 | 2.467 ± 0.106 |
5.447 ± 0.054 | 3.803 ± 0.376 |
7.091 ± 0.869 | 9.764 ± 1.448 |
5.139 ± 0.406 | 6.372 ± 1.105 |
2.467 ± 0.557 | 3.494 ± 1.229 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
