
Halovirus HHTV-2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified archaeal dsDNA viruses; Haloviruses
Average proteome isoelectric point is 4.86
Get precalculated fractions of proteins
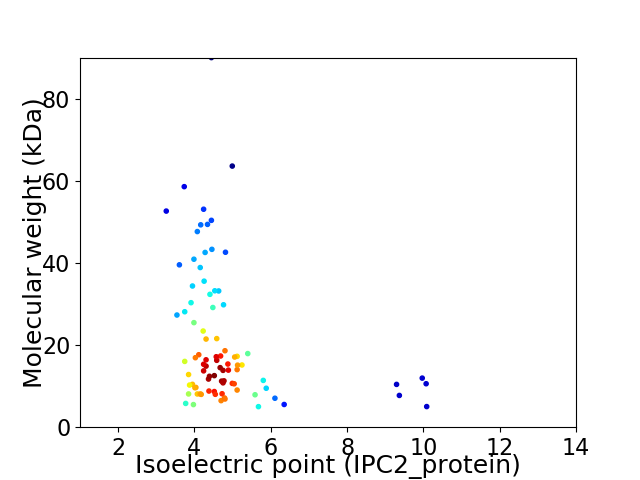
Virtual 2D-PAGE plot for 88 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4TM42|R4TM42_9VIRU Uncharacterized protein OS=Halovirus HHTV-2 OX=1273751 GN=68 PE=4 SV=1
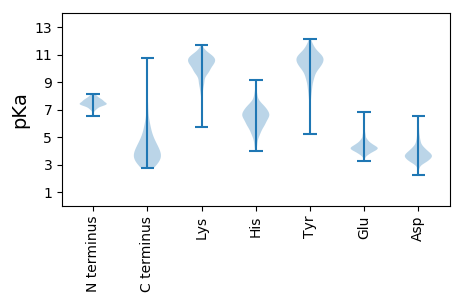
MM1 pKa = 7.85PIEE4 pKa = 4.7PIGDD8 pKa = 3.14IGIAGLTDD16 pKa = 3.45SSVTTLTWEE25 pKa = 4.26TASDD29 pKa = 3.25WDD31 pKa = 4.05SAVDD35 pKa = 3.44EE36 pKa = 5.0AGVVHH41 pKa = 6.38EE42 pKa = 5.29AVANSDD48 pKa = 3.62NDD50 pKa = 3.86DD51 pKa = 3.78ASLLKK56 pKa = 10.35QGYY59 pKa = 9.52SKK61 pKa = 9.95TSPYY65 pKa = 9.98KK66 pKa = 10.44AADD69 pKa = 4.61LIAYY73 pKa = 7.45YY74 pKa = 10.96ALDD77 pKa = 4.23EE78 pKa = 4.97DD79 pKa = 4.71SGTTVYY85 pKa = 10.88DD86 pKa = 4.11FSGNAHH92 pKa = 7.18DD93 pKa = 4.17ATNNGSTQGANGILNSTCYY112 pKa = 10.77DD113 pKa = 3.44HH114 pKa = 7.08GGSGYY119 pKa = 10.45VEE121 pKa = 5.45SDD123 pKa = 3.37DD124 pKa = 3.67QSPFGGLTAITVASWVNIGTISDD147 pKa = 3.98DD148 pKa = 4.13YY149 pKa = 11.33IVASVGDD156 pKa = 4.58LGLSDD161 pKa = 4.84ASWDD165 pKa = 3.07IWIDD169 pKa = 3.45NSGGTTGNSNTLSFDD184 pKa = 3.73MKK186 pKa = 11.1DD187 pKa = 3.39STQTNAKK194 pKa = 9.07TEE196 pKa = 4.24GSDD199 pKa = 3.28NAVPEE204 pKa = 4.58GTWALVGGTWGGGNNRR220 pKa = 11.84VWVDD224 pKa = 3.38AVDD227 pKa = 4.02VSASLEE233 pKa = 4.35GVSDD237 pKa = 3.66MQDD240 pKa = 2.8ANFPMTVGTGSDD252 pKa = 3.48HH253 pKa = 6.72TRR255 pKa = 11.84TIPDD259 pKa = 3.36RR260 pKa = 11.84QDD262 pKa = 3.5EE263 pKa = 4.56VWVWEE268 pKa = 4.53TEE270 pKa = 4.33FSQSDD275 pKa = 3.31WQTLYY280 pKa = 11.25DD281 pKa = 3.94RR282 pKa = 11.84ASTGYY287 pKa = 10.83LEE289 pKa = 4.35TATKK293 pKa = 10.58SFAASQTPDD302 pKa = 3.49LTGLDD307 pKa = 3.63YY308 pKa = 11.06TLNGVSLSVDD318 pKa = 3.77VIGSPGTGSEE328 pKa = 4.14EE329 pKa = 4.33TVTQSLTGASSYY341 pKa = 9.71TLSWSNSHH349 pKa = 5.79TDD351 pKa = 3.76FRR353 pKa = 11.84LKK355 pKa = 10.69INFDD359 pKa = 3.57TGDD362 pKa = 3.39VTTTATVNSISLSSS376 pKa = 3.24
MM1 pKa = 7.85PIEE4 pKa = 4.7PIGDD8 pKa = 3.14IGIAGLTDD16 pKa = 3.45SSVTTLTWEE25 pKa = 4.26TASDD29 pKa = 3.25WDD31 pKa = 4.05SAVDD35 pKa = 3.44EE36 pKa = 5.0AGVVHH41 pKa = 6.38EE42 pKa = 5.29AVANSDD48 pKa = 3.62NDD50 pKa = 3.86DD51 pKa = 3.78ASLLKK56 pKa = 10.35QGYY59 pKa = 9.52SKK61 pKa = 9.95TSPYY65 pKa = 9.98KK66 pKa = 10.44AADD69 pKa = 4.61LIAYY73 pKa = 7.45YY74 pKa = 10.96ALDD77 pKa = 4.23EE78 pKa = 4.97DD79 pKa = 4.71SGTTVYY85 pKa = 10.88DD86 pKa = 4.11FSGNAHH92 pKa = 7.18DD93 pKa = 4.17ATNNGSTQGANGILNSTCYY112 pKa = 10.77DD113 pKa = 3.44HH114 pKa = 7.08GGSGYY119 pKa = 10.45VEE121 pKa = 5.45SDD123 pKa = 3.37DD124 pKa = 3.67QSPFGGLTAITVASWVNIGTISDD147 pKa = 3.98DD148 pKa = 4.13YY149 pKa = 11.33IVASVGDD156 pKa = 4.58LGLSDD161 pKa = 4.84ASWDD165 pKa = 3.07IWIDD169 pKa = 3.45NSGGTTGNSNTLSFDD184 pKa = 3.73MKK186 pKa = 11.1DD187 pKa = 3.39STQTNAKK194 pKa = 9.07TEE196 pKa = 4.24GSDD199 pKa = 3.28NAVPEE204 pKa = 4.58GTWALVGGTWGGGNNRR220 pKa = 11.84VWVDD224 pKa = 3.38AVDD227 pKa = 4.02VSASLEE233 pKa = 4.35GVSDD237 pKa = 3.66MQDD240 pKa = 2.8ANFPMTVGTGSDD252 pKa = 3.48HH253 pKa = 6.72TRR255 pKa = 11.84TIPDD259 pKa = 3.36RR260 pKa = 11.84QDD262 pKa = 3.5EE263 pKa = 4.56VWVWEE268 pKa = 4.53TEE270 pKa = 4.33FSQSDD275 pKa = 3.31WQTLYY280 pKa = 11.25DD281 pKa = 3.94RR282 pKa = 11.84ASTGYY287 pKa = 10.83LEE289 pKa = 4.35TATKK293 pKa = 10.58SFAASQTPDD302 pKa = 3.49LTGLDD307 pKa = 3.63YY308 pKa = 11.06TLNGVSLSVDD318 pKa = 3.77VIGSPGTGSEE328 pKa = 4.14EE329 pKa = 4.33TVTQSLTGASSYY341 pKa = 9.71TLSWSNSHH349 pKa = 5.79TDD351 pKa = 3.76FRR353 pKa = 11.84LKK355 pKa = 10.69INFDD359 pKa = 3.57TGDD362 pKa = 3.39VTTTATVNSISLSSS376 pKa = 3.24
Molecular weight: 39.53 kDa
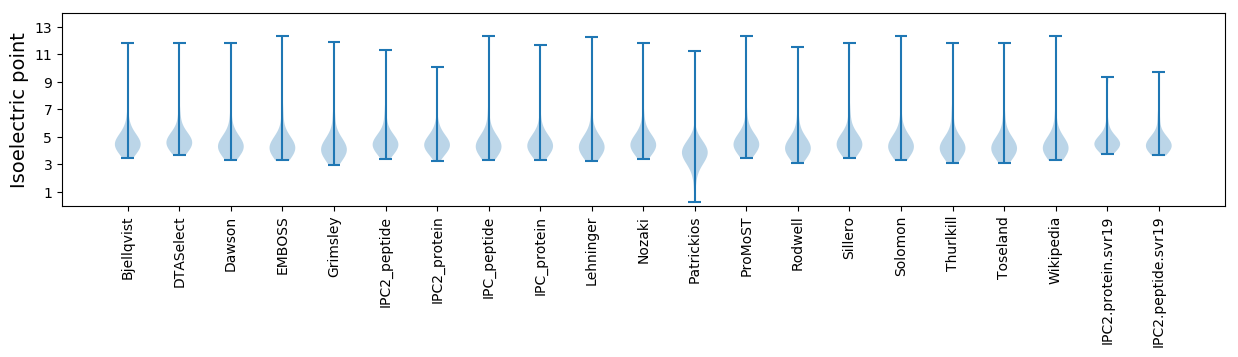
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4TG19|R4TG19_9VIRU Uncharacterized protein OS=Halovirus HHTV-2 OX=1273751 GN=4 PE=4 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84WSAARR7 pKa = 11.84RR8 pKa = 11.84DD9 pKa = 4.37AYY11 pKa = 8.41PTSYY15 pKa = 10.94PNLLLRR21 pKa = 11.84EE22 pKa = 4.36CCPVLRR28 pKa = 11.84EE29 pKa = 4.3ATPLPQGKK37 pKa = 8.41HH38 pKa = 5.23LSRR41 pKa = 11.84ATEE44 pKa = 3.87KK45 pKa = 10.85CLEE48 pKa = 4.2TEE50 pKa = 4.2EE51 pKa = 4.05PRR53 pKa = 11.84GRR55 pKa = 11.84GVPQKK60 pKa = 9.82RR61 pKa = 11.84VTRR64 pKa = 11.84IRR66 pKa = 11.84SS67 pKa = 3.27
MM1 pKa = 7.49RR2 pKa = 11.84WSAARR7 pKa = 11.84RR8 pKa = 11.84DD9 pKa = 4.37AYY11 pKa = 8.41PTSYY15 pKa = 10.94PNLLLRR21 pKa = 11.84EE22 pKa = 4.36CCPVLRR28 pKa = 11.84EE29 pKa = 4.3ATPLPQGKK37 pKa = 8.41HH38 pKa = 5.23LSRR41 pKa = 11.84ATEE44 pKa = 3.87KK45 pKa = 10.85CLEE48 pKa = 4.2TEE50 pKa = 4.2EE51 pKa = 4.05PRR53 pKa = 11.84GRR55 pKa = 11.84GVPQKK60 pKa = 9.82RR61 pKa = 11.84VTRR64 pKa = 11.84IRR66 pKa = 11.84SS67 pKa = 3.27
Molecular weight: 7.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16761 |
40 |
796 |
190.5 |
20.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.039 ± 0.294 | 0.853 ± 0.153 |
9.421 ± 0.311 | 9.576 ± 0.508 |
2.458 ± 0.16 | 8.741 ± 0.31 |
1.969 ± 0.134 | 3.275 ± 0.202 |
2.739 ± 0.207 | 6.975 ± 0.244 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.945 ± 0.116 | 2.84 ± 0.178 |
5.125 ± 0.239 | 3.371 ± 0.196 |
6.497 ± 0.371 | 6.086 ± 0.261 |
6.742 ± 0.337 | 7.613 ± 0.222 |
1.808 ± 0.125 | 2.929 ± 0.152 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
