
Grapevine virus G
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Vitivirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
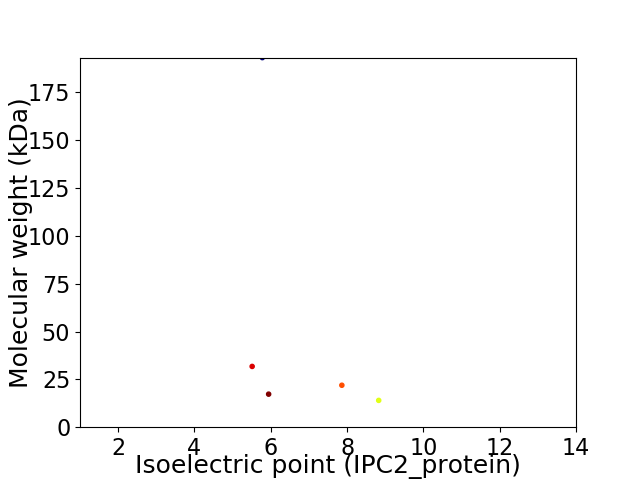
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
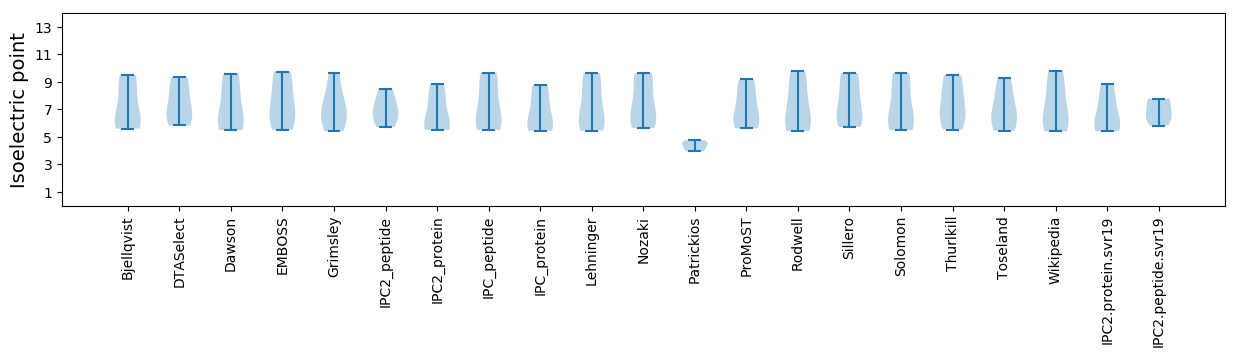
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4N967|A0A2H4N967_9VIRU Movement protein OS=Grapevine virus G OX=2022475 PE=4 SV=1
MM1 pKa = 7.3EE2 pKa = 4.65RR3 pKa = 11.84VEE5 pKa = 4.29NKK7 pKa = 9.42VFKK10 pKa = 10.79VRR12 pKa = 11.84NLKK15 pKa = 10.62NGLSNLQDD23 pKa = 3.33FEE25 pKa = 5.61KK26 pKa = 10.64IVDD29 pKa = 3.57KK30 pKa = 11.24KK31 pKa = 10.76KK32 pKa = 10.22VYY34 pKa = 10.14EE35 pKa = 4.3LGVLDD40 pKa = 5.65EE41 pKa = 4.48IFGPKK46 pKa = 9.32RR47 pKa = 11.84VFKK50 pKa = 10.79CAICKK55 pKa = 9.79EE56 pKa = 4.18LTVEE60 pKa = 4.01SGEE63 pKa = 4.52VNVQMDD69 pKa = 4.56LFCEE73 pKa = 4.09EE74 pKa = 4.42TMDD77 pKa = 5.84GIDD80 pKa = 3.58PDD82 pKa = 4.53VYY84 pKa = 10.66PFMHH88 pKa = 6.82FGCLALGFMGLGSRR102 pKa = 11.84LDD104 pKa = 3.76GQYY107 pKa = 10.28VAQVVDD113 pKa = 3.67TRR115 pKa = 11.84RR116 pKa = 11.84KK117 pKa = 8.71EE118 pKa = 3.99GSNVLSSFRR127 pKa = 11.84FRR129 pKa = 11.84CKK131 pKa = 10.52DD132 pKa = 3.16GVSAYY137 pKa = 9.77IDD139 pKa = 3.81FPDD142 pKa = 3.85FCVATTDD149 pKa = 2.94IMGGFTISIRR159 pKa = 11.84LRR161 pKa = 11.84SEE163 pKa = 5.05GIDD166 pKa = 3.66FMDD169 pKa = 5.38GSHH172 pKa = 7.3PLALNVVALCRR183 pKa = 11.84FMDD186 pKa = 4.73DD187 pKa = 3.38KK188 pKa = 11.79LEE190 pKa = 4.06TKK192 pKa = 9.92MLIKK196 pKa = 10.53KK197 pKa = 9.19HH198 pKa = 5.39GKK200 pKa = 9.74RR201 pKa = 11.84IYY203 pKa = 9.36QAVGQVEE210 pKa = 4.16ILDD213 pKa = 3.95PEE215 pKa = 4.39IGNLRR220 pKa = 11.84LNDD223 pKa = 3.97TPSGDD228 pKa = 4.21VIDD231 pKa = 3.77KK232 pKa = 10.79TMFDD236 pKa = 3.2VAEE239 pKa = 4.98IIRR242 pKa = 11.84KK243 pKa = 8.65IRR245 pKa = 11.84SSVGSGASASTKK257 pKa = 10.0DD258 pKa = 3.29GRR260 pKa = 11.84GIKK263 pKa = 9.74EE264 pKa = 4.57DD265 pKa = 3.76EE266 pKa = 4.54ASSTDD271 pKa = 3.18AAQRR275 pKa = 11.84EE276 pKa = 4.36ASGSKK281 pKa = 9.87VRR283 pKa = 11.84ALFF286 pKa = 3.77
MM1 pKa = 7.3EE2 pKa = 4.65RR3 pKa = 11.84VEE5 pKa = 4.29NKK7 pKa = 9.42VFKK10 pKa = 10.79VRR12 pKa = 11.84NLKK15 pKa = 10.62NGLSNLQDD23 pKa = 3.33FEE25 pKa = 5.61KK26 pKa = 10.64IVDD29 pKa = 3.57KK30 pKa = 11.24KK31 pKa = 10.76KK32 pKa = 10.22VYY34 pKa = 10.14EE35 pKa = 4.3LGVLDD40 pKa = 5.65EE41 pKa = 4.48IFGPKK46 pKa = 9.32RR47 pKa = 11.84VFKK50 pKa = 10.79CAICKK55 pKa = 9.79EE56 pKa = 4.18LTVEE60 pKa = 4.01SGEE63 pKa = 4.52VNVQMDD69 pKa = 4.56LFCEE73 pKa = 4.09EE74 pKa = 4.42TMDD77 pKa = 5.84GIDD80 pKa = 3.58PDD82 pKa = 4.53VYY84 pKa = 10.66PFMHH88 pKa = 6.82FGCLALGFMGLGSRR102 pKa = 11.84LDD104 pKa = 3.76GQYY107 pKa = 10.28VAQVVDD113 pKa = 3.67TRR115 pKa = 11.84RR116 pKa = 11.84KK117 pKa = 8.71EE118 pKa = 3.99GSNVLSSFRR127 pKa = 11.84FRR129 pKa = 11.84CKK131 pKa = 10.52DD132 pKa = 3.16GVSAYY137 pKa = 9.77IDD139 pKa = 3.81FPDD142 pKa = 3.85FCVATTDD149 pKa = 2.94IMGGFTISIRR159 pKa = 11.84LRR161 pKa = 11.84SEE163 pKa = 5.05GIDD166 pKa = 3.66FMDD169 pKa = 5.38GSHH172 pKa = 7.3PLALNVVALCRR183 pKa = 11.84FMDD186 pKa = 4.73DD187 pKa = 3.38KK188 pKa = 11.79LEE190 pKa = 4.06TKK192 pKa = 9.92MLIKK196 pKa = 10.53KK197 pKa = 9.19HH198 pKa = 5.39GKK200 pKa = 9.74RR201 pKa = 11.84IYY203 pKa = 9.36QAVGQVEE210 pKa = 4.16ILDD213 pKa = 3.95PEE215 pKa = 4.39IGNLRR220 pKa = 11.84LNDD223 pKa = 3.97TPSGDD228 pKa = 4.21VIDD231 pKa = 3.77KK232 pKa = 10.79TMFDD236 pKa = 3.2VAEE239 pKa = 4.98IIRR242 pKa = 11.84KK243 pKa = 8.65IRR245 pKa = 11.84SSVGSGASASTKK257 pKa = 10.0DD258 pKa = 3.29GRR260 pKa = 11.84GIKK263 pKa = 9.74EE264 pKa = 4.57DD265 pKa = 3.76EE266 pKa = 4.54ASSTDD271 pKa = 3.18AAQRR275 pKa = 11.84EE276 pKa = 4.36ASGSKK281 pKa = 9.87VRR283 pKa = 11.84ALFF286 pKa = 3.77
Molecular weight: 31.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4N957|A0A2H4N957_9VIRU Uncharacterized protein OS=Grapevine virus G OX=2022475 PE=4 SV=1
MM1 pKa = 7.05QRR3 pKa = 11.84LGEE6 pKa = 4.35SKK8 pKa = 10.38SAAKK12 pKa = 10.02RR13 pKa = 11.84RR14 pKa = 11.84AKK16 pKa = 10.38RR17 pKa = 11.84YY18 pKa = 7.65GVCYY22 pKa = 10.06KK23 pKa = 10.13CAKK26 pKa = 9.27SVCVCEE32 pKa = 4.36KK33 pKa = 10.64NRR35 pKa = 11.84ASSFDD40 pKa = 3.47RR41 pKa = 11.84TDD43 pKa = 3.1WKK45 pKa = 10.74CYY47 pKa = 8.86NAIRR51 pKa = 11.84MPATRR56 pKa = 11.84YY57 pKa = 9.79LIEE60 pKa = 4.12SRR62 pKa = 11.84GTYY65 pKa = 8.19LHH67 pKa = 6.88KK68 pKa = 10.72ASQLALSDD76 pKa = 4.05LEE78 pKa = 4.73AIGVLGYY85 pKa = 10.45EE86 pKa = 4.24KK87 pKa = 10.42YY88 pKa = 10.65VRR90 pKa = 11.84YY91 pKa = 10.12NKK93 pKa = 8.57STRR96 pKa = 11.84GCKK99 pKa = 9.95SPEE102 pKa = 3.87EE103 pKa = 4.19TPEE106 pKa = 3.49ITISINKK113 pKa = 9.61EE114 pKa = 4.07GTCLAFPLSVV124 pKa = 3.14
MM1 pKa = 7.05QRR3 pKa = 11.84LGEE6 pKa = 4.35SKK8 pKa = 10.38SAAKK12 pKa = 10.02RR13 pKa = 11.84RR14 pKa = 11.84AKK16 pKa = 10.38RR17 pKa = 11.84YY18 pKa = 7.65GVCYY22 pKa = 10.06KK23 pKa = 10.13CAKK26 pKa = 9.27SVCVCEE32 pKa = 4.36KK33 pKa = 10.64NRR35 pKa = 11.84ASSFDD40 pKa = 3.47RR41 pKa = 11.84TDD43 pKa = 3.1WKK45 pKa = 10.74CYY47 pKa = 8.86NAIRR51 pKa = 11.84MPATRR56 pKa = 11.84YY57 pKa = 9.79LIEE60 pKa = 4.12SRR62 pKa = 11.84GTYY65 pKa = 8.19LHH67 pKa = 6.88KK68 pKa = 10.72ASQLALSDD76 pKa = 4.05LEE78 pKa = 4.73AIGVLGYY85 pKa = 10.45EE86 pKa = 4.24KK87 pKa = 10.42YY88 pKa = 10.65VRR90 pKa = 11.84YY91 pKa = 10.12NKK93 pKa = 8.57STRR96 pKa = 11.84GCKK99 pKa = 9.95SPEE102 pKa = 3.87EE103 pKa = 4.19TPEE106 pKa = 3.49ITISINKK113 pKa = 9.61EE114 pKa = 4.07GTCLAFPLSVV124 pKa = 3.14
Molecular weight: 14.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2469 |
124 |
1703 |
493.8 |
55.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.197 ± 0.74 | 2.066 ± 0.411 |
5.67 ± 0.941 | 7.25 ± 0.496 |
4.901 ± 0.699 | 6.602 ± 0.517 |
2.754 ± 0.516 | 6.035 ± 0.179 |
6.926 ± 0.556 | 10.531 ± 0.987 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.714 ± 0.397 | 4.01 ± 0.483 |
3.767 ± 0.341 | 2.673 ± 0.164 |
5.306 ± 0.53 | 7.898 ± 0.702 |
4.253 ± 0.289 | 6.278 ± 0.716 |
0.527 ± 0.228 | 3.645 ± 0.555 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
