
Pan troglodytes polyomavirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
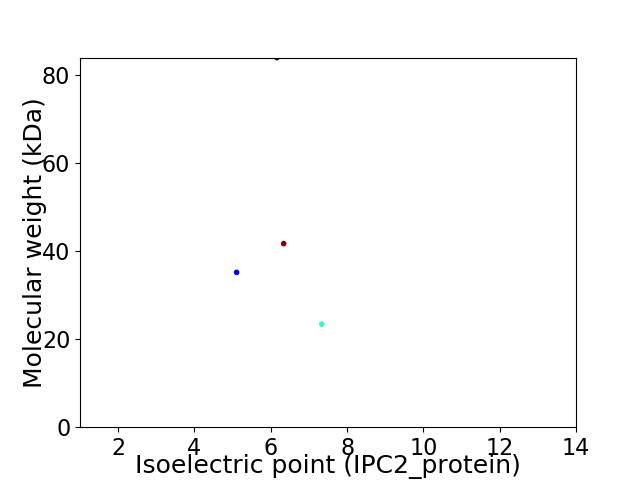
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7QK02|K7QK02_9POLY Small T antigen OS=Pan troglodytes polyomavirus 5 OX=1891737 PE=4 SV=1
MM1 pKa = 7.48GGVISIIVDD10 pKa = 4.11IIEE13 pKa = 4.14LTAEE17 pKa = 4.17LTAATGFSVEE27 pKa = 4.42AIINGEE33 pKa = 3.69ALAAVEE39 pKa = 4.4AQLSSLATVEE49 pKa = 4.17GLTGVEE55 pKa = 4.21ALASLGITTEE65 pKa = 4.02QYY67 pKa = 11.46SFLTSVPGMLSNAVGLGVVFQTVSGASALVTAGIATSYY105 pKa = 10.37AKK107 pKa = 10.13EE108 pKa = 3.83VSVVNRR114 pKa = 11.84NMAVIPWRR122 pKa = 11.84DD123 pKa = 3.0PDD125 pKa = 3.8YY126 pKa = 11.55YY127 pKa = 11.05DD128 pKa = 3.18ILFPGVQSFSYY139 pKa = 10.51AIDD142 pKa = 4.31VITDD146 pKa = 3.33WSGSLFHH153 pKa = 7.47SIGRR157 pKa = 11.84YY158 pKa = 6.41IWEE161 pKa = 4.33SVTTEE166 pKa = 3.22ARR168 pKa = 11.84RR169 pKa = 11.84QIGHH173 pKa = 7.72ASNQVIVRR181 pKa = 11.84GTHH184 pKa = 6.59AFQDD188 pKa = 3.52TLARR192 pKa = 11.84ILEE195 pKa = 3.97NARR198 pKa = 11.84WVVQSGPTNVYY209 pKa = 10.52NYY211 pKa = 10.02LDD213 pKa = 3.31NYY215 pKa = 9.21YY216 pKa = 10.54RR217 pKa = 11.84QLPPVNPAQARR228 pKa = 11.84QMYY231 pKa = 9.67RR232 pKa = 11.84RR233 pKa = 11.84LGQKK237 pKa = 10.05YY238 pKa = 8.61RR239 pKa = 11.84VTRR242 pKa = 11.84DD243 pKa = 2.96AQIPDD248 pKa = 3.76RR249 pKa = 11.84YY250 pKa = 10.07NLEE253 pKa = 3.96SSEE256 pKa = 4.37TEE258 pKa = 3.6SGEE261 pKa = 4.01TVEE264 pKa = 5.56FYY266 pKa = 10.85KK267 pKa = 10.99HH268 pKa = 6.39PGGANQRR275 pKa = 11.84VTPDD279 pKa = 2.51WMLPLILGLYY289 pKa = 10.53GDD291 pKa = 4.96ITPTWDD297 pKa = 3.24KK298 pKa = 10.89YY299 pKa = 7.19VHH301 pKa = 5.46QVEE304 pKa = 5.23EE305 pKa = 3.81EE306 pKa = 4.08DD307 pKa = 4.09EE308 pKa = 4.32YY309 pKa = 11.36QKK311 pKa = 11.12KK312 pKa = 9.61KK313 pKa = 10.08RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84VV317 pKa = 3.14
MM1 pKa = 7.48GGVISIIVDD10 pKa = 4.11IIEE13 pKa = 4.14LTAEE17 pKa = 4.17LTAATGFSVEE27 pKa = 4.42AIINGEE33 pKa = 3.69ALAAVEE39 pKa = 4.4AQLSSLATVEE49 pKa = 4.17GLTGVEE55 pKa = 4.21ALASLGITTEE65 pKa = 4.02QYY67 pKa = 11.46SFLTSVPGMLSNAVGLGVVFQTVSGASALVTAGIATSYY105 pKa = 10.37AKK107 pKa = 10.13EE108 pKa = 3.83VSVVNRR114 pKa = 11.84NMAVIPWRR122 pKa = 11.84DD123 pKa = 3.0PDD125 pKa = 3.8YY126 pKa = 11.55YY127 pKa = 11.05DD128 pKa = 3.18ILFPGVQSFSYY139 pKa = 10.51AIDD142 pKa = 4.31VITDD146 pKa = 3.33WSGSLFHH153 pKa = 7.47SIGRR157 pKa = 11.84YY158 pKa = 6.41IWEE161 pKa = 4.33SVTTEE166 pKa = 3.22ARR168 pKa = 11.84RR169 pKa = 11.84QIGHH173 pKa = 7.72ASNQVIVRR181 pKa = 11.84GTHH184 pKa = 6.59AFQDD188 pKa = 3.52TLARR192 pKa = 11.84ILEE195 pKa = 3.97NARR198 pKa = 11.84WVVQSGPTNVYY209 pKa = 10.52NYY211 pKa = 10.02LDD213 pKa = 3.31NYY215 pKa = 9.21YY216 pKa = 10.54RR217 pKa = 11.84QLPPVNPAQARR228 pKa = 11.84QMYY231 pKa = 9.67RR232 pKa = 11.84RR233 pKa = 11.84LGQKK237 pKa = 10.05YY238 pKa = 8.61RR239 pKa = 11.84VTRR242 pKa = 11.84DD243 pKa = 2.96AQIPDD248 pKa = 3.76RR249 pKa = 11.84YY250 pKa = 10.07NLEE253 pKa = 3.96SSEE256 pKa = 4.37TEE258 pKa = 3.6SGEE261 pKa = 4.01TVEE264 pKa = 5.56FYY266 pKa = 10.85KK267 pKa = 10.99HH268 pKa = 6.39PGGANQRR275 pKa = 11.84VTPDD279 pKa = 2.51WMLPLILGLYY289 pKa = 10.53GDD291 pKa = 4.96ITPTWDD297 pKa = 3.24KK298 pKa = 10.89YY299 pKa = 7.19VHH301 pKa = 5.46QVEE304 pKa = 5.23EE305 pKa = 3.81EE306 pKa = 4.08DD307 pKa = 4.09EE308 pKa = 4.32YY309 pKa = 11.36QKK311 pKa = 11.12KK312 pKa = 9.61KK313 pKa = 10.08RR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84VV317 pKa = 3.14
Molecular weight: 35.15 kDa
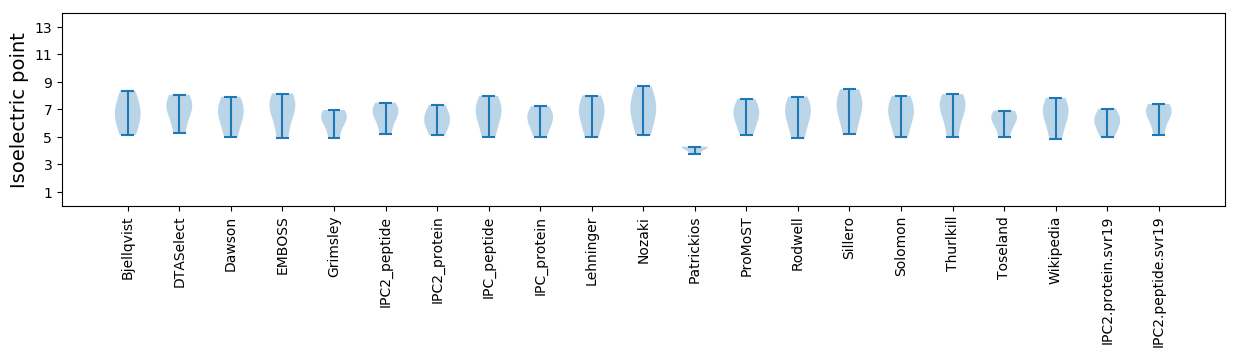
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7QKH4|K7QKH4_9POLY Large T antigen OS=Pan troglodytes polyomavirus 5 OX=1891737 PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 6.07KK3 pKa = 10.42ILHH6 pKa = 5.9KK7 pKa = 10.3SEE9 pKa = 3.97KK10 pKa = 10.05KK11 pKa = 10.3EE12 pKa = 4.08LIALLEE18 pKa = 4.3LPANEE23 pKa = 4.17YY24 pKa = 11.37GNYY27 pKa = 10.14PLMKK31 pKa = 9.9QQFKK35 pKa = 10.83KK36 pKa = 9.53MCLVYY41 pKa = 10.66HH42 pKa = 6.93PDD44 pKa = 3.32KK45 pKa = 11.54GGDD48 pKa = 3.64GEE50 pKa = 4.8KK51 pKa = 9.52MRR53 pKa = 11.84RR54 pKa = 11.84LNALWQTFNTEE65 pKa = 3.95LLEE68 pKa = 4.15MRR70 pKa = 11.84DD71 pKa = 3.36NRR73 pKa = 11.84FDD75 pKa = 3.59MEE77 pKa = 4.25QVRR80 pKa = 11.84NMDD83 pKa = 3.71LAIWEE88 pKa = 4.56DD89 pKa = 4.02LLTLKK94 pKa = 10.58EE95 pKa = 4.29VIDD98 pKa = 4.08NFDD101 pKa = 3.26EE102 pKa = 4.44VFIKK106 pKa = 9.93VFPSCFMRR114 pKa = 11.84VAVHH118 pKa = 6.03KK119 pKa = 8.59CKK121 pKa = 10.7CIVCMLRR128 pKa = 11.84QQHH131 pKa = 7.2LIMKK135 pKa = 6.63EE136 pKa = 3.87HH137 pKa = 6.45KK138 pKa = 10.13RR139 pKa = 11.84CVLWGEE145 pKa = 4.4CFCHH149 pKa = 6.77KK150 pKa = 10.44CYY152 pKa = 10.64LDD154 pKa = 3.26WYY156 pKa = 8.55GLSGIEE162 pKa = 5.17LHH164 pKa = 6.75LIWWKK169 pKa = 10.62RR170 pKa = 11.84IVQNTPFIFMRR181 pKa = 11.84VNVEE185 pKa = 3.87DD186 pKa = 5.07LYY188 pKa = 11.35HH189 pKa = 6.21KK190 pKa = 10.94RR191 pKa = 11.84NMM193 pKa = 3.91
MM1 pKa = 7.98DD2 pKa = 6.07KK3 pKa = 10.42ILHH6 pKa = 5.9KK7 pKa = 10.3SEE9 pKa = 3.97KK10 pKa = 10.05KK11 pKa = 10.3EE12 pKa = 4.08LIALLEE18 pKa = 4.3LPANEE23 pKa = 4.17YY24 pKa = 11.37GNYY27 pKa = 10.14PLMKK31 pKa = 9.9QQFKK35 pKa = 10.83KK36 pKa = 9.53MCLVYY41 pKa = 10.66HH42 pKa = 6.93PDD44 pKa = 3.32KK45 pKa = 11.54GGDD48 pKa = 3.64GEE50 pKa = 4.8KK51 pKa = 9.52MRR53 pKa = 11.84RR54 pKa = 11.84LNALWQTFNTEE65 pKa = 3.95LLEE68 pKa = 4.15MRR70 pKa = 11.84DD71 pKa = 3.36NRR73 pKa = 11.84FDD75 pKa = 3.59MEE77 pKa = 4.25QVRR80 pKa = 11.84NMDD83 pKa = 3.71LAIWEE88 pKa = 4.56DD89 pKa = 4.02LLTLKK94 pKa = 10.58EE95 pKa = 4.29VIDD98 pKa = 4.08NFDD101 pKa = 3.26EE102 pKa = 4.44VFIKK106 pKa = 9.93VFPSCFMRR114 pKa = 11.84VAVHH118 pKa = 6.03KK119 pKa = 8.59CKK121 pKa = 10.7CIVCMLRR128 pKa = 11.84QQHH131 pKa = 7.2LIMKK135 pKa = 6.63EE136 pKa = 3.87HH137 pKa = 6.45KK138 pKa = 10.13RR139 pKa = 11.84CVLWGEE145 pKa = 4.4CFCHH149 pKa = 6.77KK150 pKa = 10.44CYY152 pKa = 10.64LDD154 pKa = 3.26WYY156 pKa = 8.55GLSGIEE162 pKa = 5.17LHH164 pKa = 6.75LIWWKK169 pKa = 10.62RR170 pKa = 11.84IVQNTPFIFMRR181 pKa = 11.84VNVEE185 pKa = 3.87DD186 pKa = 5.07LYY188 pKa = 11.35HH189 pKa = 6.21KK190 pKa = 10.94RR191 pKa = 11.84NMM193 pKa = 3.91
Molecular weight: 23.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1626 |
193 |
733 |
406.5 |
46.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.781 ± 0.741 | 2.583 ± 0.624 |
5.535 ± 0.295 | 6.704 ± 0.357 |
4.49 ± 0.682 | 5.535 ± 0.936 |
1.845 ± 0.362 | 5.105 ± 0.469 |
7.134 ± 1.199 | 9.102 ± 0.549 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.014 ± 0.48 | 4.92 ± 0.295 |
5.474 ± 0.674 | 4.059 ± 0.291 |
4.551 ± 0.453 | 7.011 ± 0.839 |
5.843 ± 0.903 | 6.396 ± 1.161 |
1.292 ± 0.353 | 3.629 ± 0.439 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
