
Beihai anemone virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
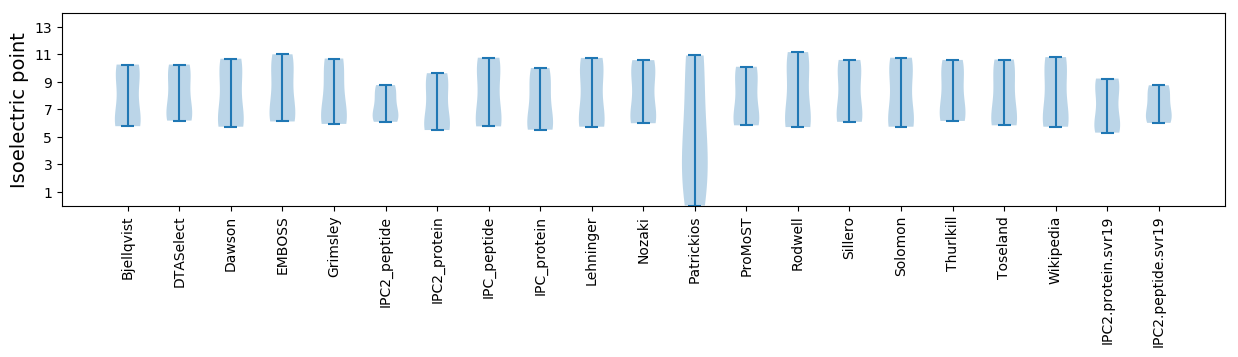
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins
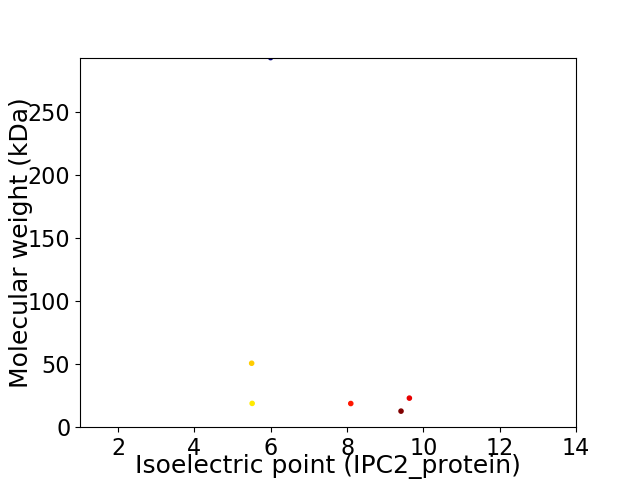
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJG1|A0A1L3KJG1_9VIRU Uncharacterized protein OS=Beihai anemone virus 1 OX=1922352 PE=4 SV=1
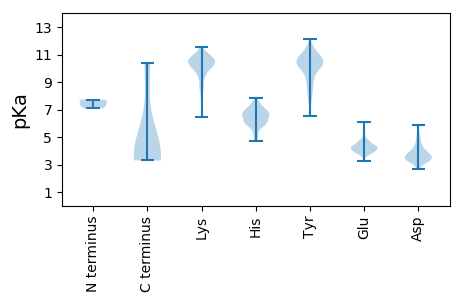
MM1 pKa = 7.72PFYY4 pKa = 10.34MDD6 pKa = 3.81FRR8 pKa = 11.84LYY10 pKa = 10.92SPLVYY15 pKa = 10.4LVYY18 pKa = 10.54LYY20 pKa = 11.32VSLICSLSASSVAVAEE36 pKa = 5.04PILTSSQALLLDD48 pKa = 3.25YY49 pKa = 10.94DD50 pKa = 4.31YY51 pKa = 11.46FNKK54 pKa = 10.32LSSLLYY60 pKa = 10.14YY61 pKa = 10.42RR62 pKa = 11.84SEE64 pKa = 3.81LLIYY68 pKa = 9.46PFKK71 pKa = 10.84TIDD74 pKa = 3.61FQCATTYY81 pKa = 10.7EE82 pKa = 4.01IGNLDD87 pKa = 3.3FHH89 pKa = 6.84EE90 pKa = 4.73CVLPFSCGNFVEE102 pKa = 4.33NQLIEE107 pKa = 4.32GGYY110 pKa = 10.0FSSDD114 pKa = 3.43YY115 pKa = 10.95LCYY118 pKa = 10.07RR119 pKa = 11.84YY120 pKa = 9.9AYY122 pKa = 10.05EE123 pKa = 4.66GDD125 pKa = 3.07TRR127 pKa = 11.84IRR129 pKa = 11.84QFNNSLSTVKK139 pKa = 10.68LGVFGKK145 pKa = 9.95FLISTHH151 pKa = 5.99HH152 pKa = 6.36FPVIVEE158 pKa = 4.22SPRR161 pKa = 11.84GEE163 pKa = 4.34LFLSDD168 pKa = 4.69FRR170 pKa = 11.84SLDD173 pKa = 3.21LYY175 pKa = 11.28ARR177 pKa = 11.84SDD179 pKa = 3.55LNSRR183 pKa = 11.84FTASNFRR190 pKa = 11.84PDD192 pKa = 4.54CIRR195 pKa = 11.84TNITQILFPVDD206 pKa = 3.17PLGPVNIYY214 pKa = 10.52QYY216 pKa = 11.29LNHH219 pKa = 6.93ILNTRR224 pKa = 11.84FTIEE228 pKa = 3.84ILPLYY233 pKa = 10.44SEE235 pKa = 6.0LIYY238 pKa = 10.88FPLNKK243 pKa = 9.32PDD245 pKa = 3.35ILTSVRR251 pKa = 11.84FTGTFQIDD259 pKa = 4.24CQTTIYY265 pKa = 9.88TFSNVTIRR273 pKa = 11.84YY274 pKa = 7.27KK275 pKa = 11.04KK276 pKa = 10.18NVSNSDD282 pKa = 3.24ILDD285 pKa = 4.02LEE287 pKa = 4.7VVGQGGGKK295 pKa = 9.91FPVRR299 pKa = 11.84DD300 pKa = 3.93CHH302 pKa = 6.84CVPVDD307 pKa = 3.54SAHH310 pKa = 6.85FLIEE314 pKa = 4.1YY315 pKa = 9.8GDD317 pKa = 3.88PFAINHH323 pKa = 5.26TLINVTEE330 pKa = 4.16KK331 pKa = 10.98CVVATYY337 pKa = 10.42KK338 pKa = 10.93RR339 pKa = 11.84NDD341 pKa = 3.02HH342 pKa = 6.3SFWQIIVHH350 pKa = 6.37DD351 pKa = 5.11FISSLRR357 pKa = 11.84GLIYY361 pKa = 10.67DD362 pKa = 5.21FIDD365 pKa = 2.89IVKK368 pKa = 10.15FSLSDD373 pKa = 3.34FVKK376 pKa = 10.36QFRR379 pKa = 11.84DD380 pKa = 4.93FILQSFKK387 pKa = 10.88TLIEE391 pKa = 4.08YY392 pKa = 10.3LFHH395 pKa = 7.31LEE397 pKa = 3.99RR398 pKa = 11.84EE399 pKa = 4.09IKK401 pKa = 10.2IIEE404 pKa = 4.08FFIIFLVLINKK415 pKa = 7.7FQHH418 pKa = 6.46PLSACIVAVIVWILTPFSTKK438 pKa = 10.36
MM1 pKa = 7.72PFYY4 pKa = 10.34MDD6 pKa = 3.81FRR8 pKa = 11.84LYY10 pKa = 10.92SPLVYY15 pKa = 10.4LVYY18 pKa = 10.54LYY20 pKa = 11.32VSLICSLSASSVAVAEE36 pKa = 5.04PILTSSQALLLDD48 pKa = 3.25YY49 pKa = 10.94DD50 pKa = 4.31YY51 pKa = 11.46FNKK54 pKa = 10.32LSSLLYY60 pKa = 10.14YY61 pKa = 10.42RR62 pKa = 11.84SEE64 pKa = 3.81LLIYY68 pKa = 9.46PFKK71 pKa = 10.84TIDD74 pKa = 3.61FQCATTYY81 pKa = 10.7EE82 pKa = 4.01IGNLDD87 pKa = 3.3FHH89 pKa = 6.84EE90 pKa = 4.73CVLPFSCGNFVEE102 pKa = 4.33NQLIEE107 pKa = 4.32GGYY110 pKa = 10.0FSSDD114 pKa = 3.43YY115 pKa = 10.95LCYY118 pKa = 10.07RR119 pKa = 11.84YY120 pKa = 9.9AYY122 pKa = 10.05EE123 pKa = 4.66GDD125 pKa = 3.07TRR127 pKa = 11.84IRR129 pKa = 11.84QFNNSLSTVKK139 pKa = 10.68LGVFGKK145 pKa = 9.95FLISTHH151 pKa = 5.99HH152 pKa = 6.36FPVIVEE158 pKa = 4.22SPRR161 pKa = 11.84GEE163 pKa = 4.34LFLSDD168 pKa = 4.69FRR170 pKa = 11.84SLDD173 pKa = 3.21LYY175 pKa = 11.28ARR177 pKa = 11.84SDD179 pKa = 3.55LNSRR183 pKa = 11.84FTASNFRR190 pKa = 11.84PDD192 pKa = 4.54CIRR195 pKa = 11.84TNITQILFPVDD206 pKa = 3.17PLGPVNIYY214 pKa = 10.52QYY216 pKa = 11.29LNHH219 pKa = 6.93ILNTRR224 pKa = 11.84FTIEE228 pKa = 3.84ILPLYY233 pKa = 10.44SEE235 pKa = 6.0LIYY238 pKa = 10.88FPLNKK243 pKa = 9.32PDD245 pKa = 3.35ILTSVRR251 pKa = 11.84FTGTFQIDD259 pKa = 4.24CQTTIYY265 pKa = 9.88TFSNVTIRR273 pKa = 11.84YY274 pKa = 7.27KK275 pKa = 11.04KK276 pKa = 10.18NVSNSDD282 pKa = 3.24ILDD285 pKa = 4.02LEE287 pKa = 4.7VVGQGGGKK295 pKa = 9.91FPVRR299 pKa = 11.84DD300 pKa = 3.93CHH302 pKa = 6.84CVPVDD307 pKa = 3.54SAHH310 pKa = 6.85FLIEE314 pKa = 4.1YY315 pKa = 9.8GDD317 pKa = 3.88PFAINHH323 pKa = 5.26TLINVTEE330 pKa = 4.16KK331 pKa = 10.98CVVATYY337 pKa = 10.42KK338 pKa = 10.93RR339 pKa = 11.84NDD341 pKa = 3.02HH342 pKa = 6.3SFWQIIVHH350 pKa = 6.37DD351 pKa = 5.11FISSLRR357 pKa = 11.84GLIYY361 pKa = 10.67DD362 pKa = 5.21FIDD365 pKa = 2.89IVKK368 pKa = 10.15FSLSDD373 pKa = 3.34FVKK376 pKa = 10.36QFRR379 pKa = 11.84DD380 pKa = 4.93FILQSFKK387 pKa = 10.88TLIEE391 pKa = 4.08YY392 pKa = 10.3LFHH395 pKa = 7.31LEE397 pKa = 3.99RR398 pKa = 11.84EE399 pKa = 4.09IKK401 pKa = 10.2IIEE404 pKa = 4.08FFIIFLVLINKK415 pKa = 7.7FQHH418 pKa = 6.46PLSACIVAVIVWILTPFSTKK438 pKa = 10.36
Molecular weight: 50.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJR7|A0A1L3KJR7_9VIRU Uncharacterized protein OS=Beihai anemone virus 1 OX=1922352 PE=4 SV=1
MM1 pKa = 7.24MNTVRR6 pKa = 11.84RR7 pKa = 11.84INGRR11 pKa = 11.84ALSRR15 pKa = 11.84SPPIIFDD22 pKa = 3.77YY23 pKa = 10.64PPPLPQRR30 pKa = 11.84FKK32 pKa = 10.79PPPAPKK38 pKa = 9.73PKK40 pKa = 10.08RR41 pKa = 11.84SDD43 pKa = 3.25VFTEE47 pKa = 4.08LMDD50 pKa = 4.86GIKK53 pKa = 10.34KK54 pKa = 8.15ITQQPDD60 pKa = 3.12VLALLLLALIIVYY73 pKa = 9.99DD74 pKa = 3.8ISHH77 pKa = 7.74LDD79 pKa = 3.29RR80 pKa = 11.84LASFLGRR87 pKa = 11.84KK88 pKa = 8.21EE89 pKa = 3.91AVKK92 pKa = 10.4PFADD96 pKa = 4.1WISSNKK102 pKa = 7.4TQIAGLIILFPALYY116 pKa = 10.05KK117 pKa = 10.53FPQRR121 pKa = 11.84SRR123 pKa = 11.84WIAFGVAFLTIFAMAPQNLLVYY145 pKa = 9.29LFISAGILLYY155 pKa = 9.74WQIPRR160 pKa = 11.84PEE162 pKa = 3.84FRR164 pKa = 11.84LFLLIVAGVYY174 pKa = 9.42FYY176 pKa = 10.84ILFVEE181 pKa = 4.79TVPDD185 pKa = 4.33PVPVTPPNPVPVVEE199 pKa = 4.21RR200 pKa = 11.84RR201 pKa = 11.84KK202 pKa = 10.56GG203 pKa = 3.3
MM1 pKa = 7.24MNTVRR6 pKa = 11.84RR7 pKa = 11.84INGRR11 pKa = 11.84ALSRR15 pKa = 11.84SPPIIFDD22 pKa = 3.77YY23 pKa = 10.64PPPLPQRR30 pKa = 11.84FKK32 pKa = 10.79PPPAPKK38 pKa = 9.73PKK40 pKa = 10.08RR41 pKa = 11.84SDD43 pKa = 3.25VFTEE47 pKa = 4.08LMDD50 pKa = 4.86GIKK53 pKa = 10.34KK54 pKa = 8.15ITQQPDD60 pKa = 3.12VLALLLLALIIVYY73 pKa = 9.99DD74 pKa = 3.8ISHH77 pKa = 7.74LDD79 pKa = 3.29RR80 pKa = 11.84LASFLGRR87 pKa = 11.84KK88 pKa = 8.21EE89 pKa = 3.91AVKK92 pKa = 10.4PFADD96 pKa = 4.1WISSNKK102 pKa = 7.4TQIAGLIILFPALYY116 pKa = 10.05KK117 pKa = 10.53FPQRR121 pKa = 11.84SRR123 pKa = 11.84WIAFGVAFLTIFAMAPQNLLVYY145 pKa = 9.29LFISAGILLYY155 pKa = 9.74WQIPRR160 pKa = 11.84PEE162 pKa = 3.84FRR164 pKa = 11.84LFLLIVAGVYY174 pKa = 9.42FYY176 pKa = 10.84ILFVEE181 pKa = 4.79TVPDD185 pKa = 4.33PVPVTPPNPVPVVEE199 pKa = 4.21RR200 pKa = 11.84RR201 pKa = 11.84KK202 pKa = 10.56GG203 pKa = 3.3
Molecular weight: 23.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3681 |
110 |
2601 |
613.5 |
69.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.488 ± 0.845 | 2.282 ± 0.714 |
6.52 ± 0.945 | 5.053 ± 0.568 |
5.868 ± 1.128 | 4.319 ± 0.339 |
2.526 ± 0.384 | 7.281 ± 0.622 |
6.982 ± 1.238 | 9.182 ± 1.195 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.657 ± 0.358 | 5.352 ± 0.678 |
4.863 ± 1.273 | 2.662 ± 0.532 |
4.319 ± 0.861 | 7.688 ± 0.548 |
5.379 ± 0.535 | 7.634 ± 0.46 |
0.625 ± 0.172 | 4.319 ± 0.768 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
