
Salmonella phage vB_SenM_PA13076
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Rosemountvirus; unclassified Rosemountvirus
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
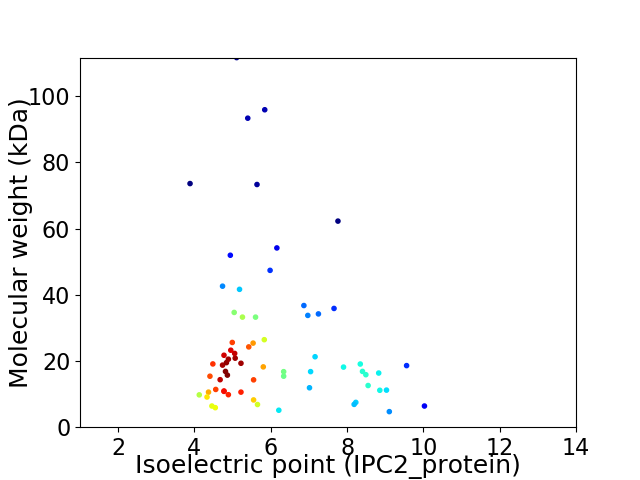
Virtual 2D-PAGE plot for 68 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z2U3U7|A0A2Z2U3U7_9CAUD Scaffold protein OS=Salmonella phage vB_SenM_PA13076 OX=2029636 GN=PA13076_16 PE=4 SV=1
MM1 pKa = 7.6SLPLNEE7 pKa = 4.29VVDD10 pKa = 4.24VQFSKK15 pKa = 11.57SNGGKK20 pKa = 10.28GFTPNVPLEE29 pKa = 4.49GIMLSNYY36 pKa = 9.14EE37 pKa = 3.98YY38 pKa = 10.99SIVAGDD44 pKa = 4.53DD45 pKa = 3.36PVQLVVRR52 pKa = 11.84YY53 pKa = 9.45IPTNATEE60 pKa = 5.76RR61 pKa = 11.84GMAWKK66 pKa = 10.58SSDD69 pKa = 3.54PDD71 pKa = 3.86IITVDD76 pKa = 3.11EE77 pKa = 4.67LGFVKK82 pKa = 10.41PLAEE86 pKa = 3.89GVAYY90 pKa = 8.99VTGTSVDD97 pKa = 2.63GGYY100 pKa = 7.85TVRR103 pKa = 11.84AKK105 pKa = 10.76FNIARR110 pKa = 11.84APILVEE116 pKa = 4.12SVTVSPTSVTLEE128 pKa = 3.79EE129 pKa = 5.05GKK131 pKa = 7.58TQRR134 pKa = 11.84LTYY137 pKa = 9.93TYY139 pKa = 10.92LPANADD145 pKa = 3.32KK146 pKa = 10.92PDD148 pKa = 4.42FYY150 pKa = 10.54WISDD154 pKa = 3.91DD155 pKa = 3.74EE156 pKa = 4.67SVAVVNDD163 pKa = 3.71SGLVTAIGDD172 pKa = 3.75GEE174 pKa = 4.59TTVNVQYY181 pKa = 11.0GVDD184 pKa = 3.41SSIFDD189 pKa = 3.6SCTVTVIPPVVHH201 pKa = 4.41VTGVEE206 pKa = 4.19FLTQSPLNVNIGSTAGTSIRR226 pKa = 11.84ISPEE230 pKa = 3.42DD231 pKa = 3.54ATDD234 pKa = 3.29KK235 pKa = 10.38TVTYY239 pKa = 9.31EE240 pKa = 4.0SSAPTVATVDD250 pKa = 2.91TDD252 pKa = 4.0GNVTGIIAGNATVTVKK268 pKa = 10.27TNDD271 pKa = 3.15GDD273 pKa = 3.81KK274 pKa = 10.88TDD276 pKa = 3.67TLDD279 pKa = 3.83IIVRR283 pKa = 11.84VPAVNVTGVSIDD295 pKa = 3.81EE296 pKa = 4.66GDD298 pKa = 3.67TASMEE303 pKa = 4.42VGDD306 pKa = 4.33TLQLHH311 pKa = 5.73ATVTPEE317 pKa = 3.61NATDD321 pKa = 4.1PSVVWSAPVSEE332 pKa = 4.44VASVTEE338 pKa = 4.29AGLVTANAAGLIDD351 pKa = 3.69VTVITNDD358 pKa = 2.97GNFTDD363 pKa = 4.8RR364 pKa = 11.84IEE366 pKa = 4.53VKK368 pKa = 9.66VTDD371 pKa = 3.8PRR373 pKa = 11.84IAVTDD378 pKa = 3.9VAWATDD384 pKa = 3.46SPTKK388 pKa = 10.56VSAGHH393 pKa = 5.14QVQTKK398 pKa = 10.14IVITPDD404 pKa = 2.63NATNKK409 pKa = 9.3KK410 pKa = 10.01VSYY413 pKa = 10.43NSSNTDD419 pKa = 2.75AVTVDD424 pKa = 3.83TNGVIYY430 pKa = 9.67GQSPGTSTITVTTDD444 pKa = 2.91DD445 pKa = 4.25GGLVASFVATCTEE458 pKa = 4.2VAVITPDD465 pKa = 3.35GLGGIPVGGTQQFTYY480 pKa = 9.78TVNPSDD486 pKa = 5.01ADD488 pKa = 3.63LTNIRR493 pKa = 11.84YY494 pKa = 8.83EE495 pKa = 4.22SANPDD500 pKa = 3.1IATVDD505 pKa = 3.28SSGVVTGVASGGCNIWIYY523 pKa = 9.84ATSNGADD530 pKa = 2.96ISAATWAQISEE541 pKa = 4.36KK542 pKa = 10.0VQVYY546 pKa = 8.38PQYY549 pKa = 11.5AEE551 pKa = 3.83WLVGNTKK558 pKa = 10.19QLDD561 pKa = 3.7VEE563 pKa = 4.89IYY565 pKa = 9.81PSDD568 pKa = 3.82SKK570 pKa = 11.51SVTVTYY576 pKa = 10.2EE577 pKa = 3.74SSNDD581 pKa = 3.65AIISIDD587 pKa = 3.57ANGLMTAKK595 pKa = 10.34SEE597 pKa = 4.15GSATITANVVMNGVAASGTTSPYY620 pKa = 10.0VSEE623 pKa = 5.1LSVTTDD629 pKa = 3.11SLPALTVGNTQQLVVTVRR647 pKa = 11.84PEE649 pKa = 3.78YY650 pKa = 11.1AKK652 pKa = 9.91TDD654 pKa = 3.48PGYY657 pKa = 10.56NIQYY661 pKa = 7.06TTTDD665 pKa = 3.4DD666 pKa = 3.49TVATVSNTGVITGVGDD682 pKa = 3.7GGCRR686 pKa = 11.84IGATVTVRR694 pKa = 11.84NATASDD700 pKa = 3.35SSYY703 pKa = 11.09QSVDD707 pKa = 2.9AAA709 pKa = 4.15
MM1 pKa = 7.6SLPLNEE7 pKa = 4.29VVDD10 pKa = 4.24VQFSKK15 pKa = 11.57SNGGKK20 pKa = 10.28GFTPNVPLEE29 pKa = 4.49GIMLSNYY36 pKa = 9.14EE37 pKa = 3.98YY38 pKa = 10.99SIVAGDD44 pKa = 4.53DD45 pKa = 3.36PVQLVVRR52 pKa = 11.84YY53 pKa = 9.45IPTNATEE60 pKa = 5.76RR61 pKa = 11.84GMAWKK66 pKa = 10.58SSDD69 pKa = 3.54PDD71 pKa = 3.86IITVDD76 pKa = 3.11EE77 pKa = 4.67LGFVKK82 pKa = 10.41PLAEE86 pKa = 3.89GVAYY90 pKa = 8.99VTGTSVDD97 pKa = 2.63GGYY100 pKa = 7.85TVRR103 pKa = 11.84AKK105 pKa = 10.76FNIARR110 pKa = 11.84APILVEE116 pKa = 4.12SVTVSPTSVTLEE128 pKa = 3.79EE129 pKa = 5.05GKK131 pKa = 7.58TQRR134 pKa = 11.84LTYY137 pKa = 9.93TYY139 pKa = 10.92LPANADD145 pKa = 3.32KK146 pKa = 10.92PDD148 pKa = 4.42FYY150 pKa = 10.54WISDD154 pKa = 3.91DD155 pKa = 3.74EE156 pKa = 4.67SVAVVNDD163 pKa = 3.71SGLVTAIGDD172 pKa = 3.75GEE174 pKa = 4.59TTVNVQYY181 pKa = 11.0GVDD184 pKa = 3.41SSIFDD189 pKa = 3.6SCTVTVIPPVVHH201 pKa = 4.41VTGVEE206 pKa = 4.19FLTQSPLNVNIGSTAGTSIRR226 pKa = 11.84ISPEE230 pKa = 3.42DD231 pKa = 3.54ATDD234 pKa = 3.29KK235 pKa = 10.38TVTYY239 pKa = 9.31EE240 pKa = 4.0SSAPTVATVDD250 pKa = 2.91TDD252 pKa = 4.0GNVTGIIAGNATVTVKK268 pKa = 10.27TNDD271 pKa = 3.15GDD273 pKa = 3.81KK274 pKa = 10.88TDD276 pKa = 3.67TLDD279 pKa = 3.83IIVRR283 pKa = 11.84VPAVNVTGVSIDD295 pKa = 3.81EE296 pKa = 4.66GDD298 pKa = 3.67TASMEE303 pKa = 4.42VGDD306 pKa = 4.33TLQLHH311 pKa = 5.73ATVTPEE317 pKa = 3.61NATDD321 pKa = 4.1PSVVWSAPVSEE332 pKa = 4.44VASVTEE338 pKa = 4.29AGLVTANAAGLIDD351 pKa = 3.69VTVITNDD358 pKa = 2.97GNFTDD363 pKa = 4.8RR364 pKa = 11.84IEE366 pKa = 4.53VKK368 pKa = 9.66VTDD371 pKa = 3.8PRR373 pKa = 11.84IAVTDD378 pKa = 3.9VAWATDD384 pKa = 3.46SPTKK388 pKa = 10.56VSAGHH393 pKa = 5.14QVQTKK398 pKa = 10.14IVITPDD404 pKa = 2.63NATNKK409 pKa = 9.3KK410 pKa = 10.01VSYY413 pKa = 10.43NSSNTDD419 pKa = 2.75AVTVDD424 pKa = 3.83TNGVIYY430 pKa = 9.67GQSPGTSTITVTTDD444 pKa = 2.91DD445 pKa = 4.25GGLVASFVATCTEE458 pKa = 4.2VAVITPDD465 pKa = 3.35GLGGIPVGGTQQFTYY480 pKa = 9.78TVNPSDD486 pKa = 5.01ADD488 pKa = 3.63LTNIRR493 pKa = 11.84YY494 pKa = 8.83EE495 pKa = 4.22SANPDD500 pKa = 3.1IATVDD505 pKa = 3.28SSGVVTGVASGGCNIWIYY523 pKa = 9.84ATSNGADD530 pKa = 2.96ISAATWAQISEE541 pKa = 4.36KK542 pKa = 10.0VQVYY546 pKa = 8.38PQYY549 pKa = 11.5AEE551 pKa = 3.83WLVGNTKK558 pKa = 10.19QLDD561 pKa = 3.7VEE563 pKa = 4.89IYY565 pKa = 9.81PSDD568 pKa = 3.82SKK570 pKa = 11.51SVTVTYY576 pKa = 10.2EE577 pKa = 3.74SSNDD581 pKa = 3.65AIISIDD587 pKa = 3.57ANGLMTAKK595 pKa = 10.34SEE597 pKa = 4.15GSATITANVVMNGVAASGTTSPYY620 pKa = 10.0VSEE623 pKa = 5.1LSVTTDD629 pKa = 3.11SLPALTVGNTQQLVVTVRR647 pKa = 11.84PEE649 pKa = 3.78YY650 pKa = 11.1AKK652 pKa = 9.91TDD654 pKa = 3.48PGYY657 pKa = 10.56NIQYY661 pKa = 7.06TTTDD665 pKa = 3.4DD666 pKa = 3.49TVATVSNTGVITGVGDD682 pKa = 3.7GGCRR686 pKa = 11.84IGATVTVRR694 pKa = 11.84NATASDD700 pKa = 3.35SSYY703 pKa = 11.09QSVDD707 pKa = 2.9AAA709 pKa = 4.15
Molecular weight: 73.63 kDa
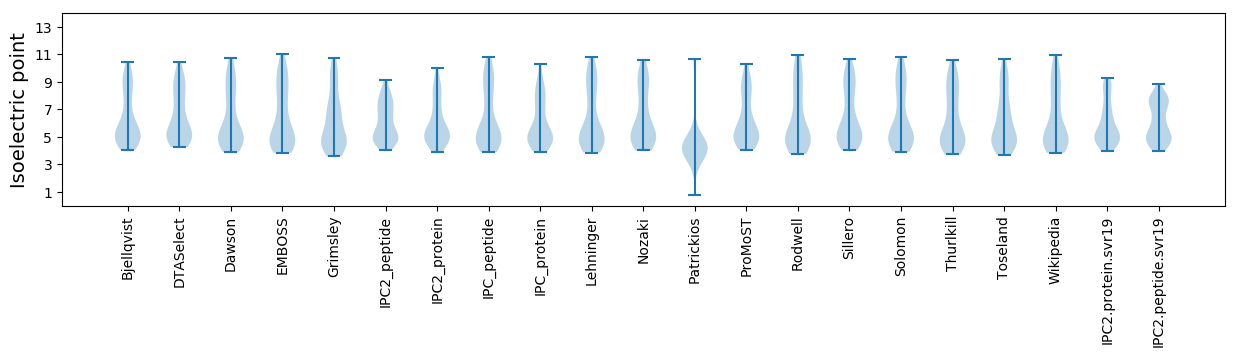
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z2U5W7|A0A2Z2U5W7_9CAUD Uncharacterized protein OS=Salmonella phage vB_SenM_PA13076 OX=2029636 GN=PA13076_23 PE=4 SV=1
MM1 pKa = 7.46IRR3 pKa = 11.84IRR5 pKa = 11.84LEE7 pKa = 3.74RR8 pKa = 11.84RR9 pKa = 11.84DD10 pKa = 3.51KK11 pKa = 11.14QKK13 pKa = 11.29FEE15 pKa = 3.54FTYY18 pKa = 10.47RR19 pKa = 11.84AFKK22 pKa = 10.7SFEE25 pKa = 4.05GQGVSVGVHH34 pKa = 6.16PEE36 pKa = 3.42HH37 pKa = 6.82NRR39 pKa = 11.84RR40 pKa = 11.84INPRR44 pKa = 11.84DD45 pKa = 3.53RR46 pKa = 11.84ITNAEE51 pKa = 3.92LAMVHH56 pKa = 6.6EE57 pKa = 4.93YY58 pKa = 11.07GLNGLPEE65 pKa = 4.16RR66 pKa = 11.84SFLRR70 pKa = 11.84SAVGGRR76 pKa = 11.84GKK78 pKa = 10.56GRR80 pKa = 11.84QAINKK85 pKa = 8.43AFRR88 pKa = 11.84DD89 pKa = 3.76NVPAVLRR96 pKa = 11.84GSMTAHH102 pKa = 7.53KK103 pKa = 10.47LNDD106 pKa = 3.85KK107 pKa = 9.78IGRR110 pKa = 11.84MLVDD114 pKa = 4.0AVHH117 pKa = 7.13DD118 pKa = 4.29RR119 pKa = 11.84MDD121 pKa = 4.21SDD123 pKa = 4.09VPPPNTEE130 pKa = 3.51LTEE133 pKa = 3.94EE134 pKa = 4.36RR135 pKa = 11.84KK136 pKa = 10.02RR137 pKa = 11.84GPGTLRR143 pKa = 11.84EE144 pKa = 4.05SMQLYY149 pKa = 10.63DD150 pKa = 3.76SIGYY154 pKa = 9.31KK155 pKa = 9.94VGRR158 pKa = 11.84NWRR161 pKa = 11.84LL162 pKa = 2.91
MM1 pKa = 7.46IRR3 pKa = 11.84IRR5 pKa = 11.84LEE7 pKa = 3.74RR8 pKa = 11.84RR9 pKa = 11.84DD10 pKa = 3.51KK11 pKa = 11.14QKK13 pKa = 11.29FEE15 pKa = 3.54FTYY18 pKa = 10.47RR19 pKa = 11.84AFKK22 pKa = 10.7SFEE25 pKa = 4.05GQGVSVGVHH34 pKa = 6.16PEE36 pKa = 3.42HH37 pKa = 6.82NRR39 pKa = 11.84RR40 pKa = 11.84INPRR44 pKa = 11.84DD45 pKa = 3.53RR46 pKa = 11.84ITNAEE51 pKa = 3.92LAMVHH56 pKa = 6.6EE57 pKa = 4.93YY58 pKa = 11.07GLNGLPEE65 pKa = 4.16RR66 pKa = 11.84SFLRR70 pKa = 11.84SAVGGRR76 pKa = 11.84GKK78 pKa = 10.56GRR80 pKa = 11.84QAINKK85 pKa = 8.43AFRR88 pKa = 11.84DD89 pKa = 3.76NVPAVLRR96 pKa = 11.84GSMTAHH102 pKa = 7.53KK103 pKa = 10.47LNDD106 pKa = 3.85KK107 pKa = 9.78IGRR110 pKa = 11.84MLVDD114 pKa = 4.0AVHH117 pKa = 7.13DD118 pKa = 4.29RR119 pKa = 11.84MDD121 pKa = 4.21SDD123 pKa = 4.09VPPPNTEE130 pKa = 3.51LTEE133 pKa = 3.94EE134 pKa = 4.36RR135 pKa = 11.84KK136 pKa = 10.02RR137 pKa = 11.84GPGTLRR143 pKa = 11.84EE144 pKa = 4.05SMQLYY149 pKa = 10.63DD150 pKa = 3.76SIGYY154 pKa = 9.31KK155 pKa = 9.94VGRR158 pKa = 11.84NWRR161 pKa = 11.84LL162 pKa = 2.91
Molecular weight: 18.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15560 |
41 |
985 |
228.8 |
25.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.393 ± 0.306 | 1.131 ± 0.111 |
6.414 ± 0.176 | 6.298 ± 0.338 |
3.445 ± 0.175 | 6.979 ± 0.276 |
1.69 ± 0.159 | 5.9 ± 0.177 |
6.138 ± 0.332 | 7.808 ± 0.216 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.808 ± 0.178 | 5.289 ± 0.178 |
4.165 ± 0.24 | 3.766 ± 0.182 |
4.679 ± 0.206 | 5.649 ± 0.27 |
6.433 ± 0.415 | 7.635 ± 0.397 |
1.684 ± 0.152 | 3.695 ± 0.253 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
