
Hubei sobemo-like virus 38
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
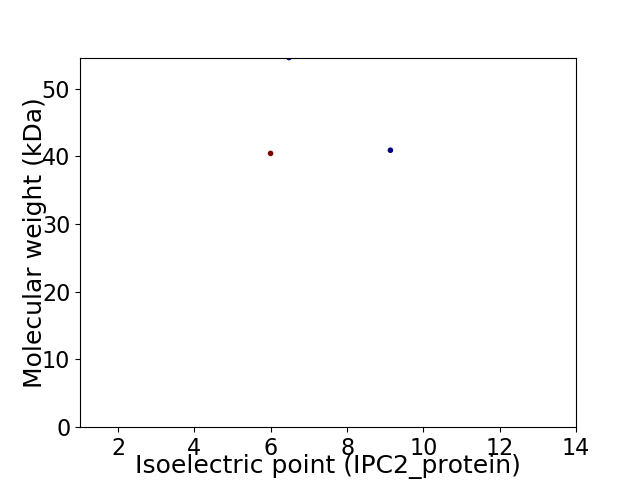
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEJ2|A0A1L3KEJ2_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 38 OX=1923225 PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 6.19ASPGLPFCKK11 pKa = 9.71HH12 pKa = 5.33YY13 pKa = 9.37KK14 pKa = 9.39TNAEE18 pKa = 4.09LLGFDD23 pKa = 5.18GFNCDD28 pKa = 4.91AVKK31 pKa = 10.64ADD33 pKa = 3.73CLVNMFYY40 pKa = 11.17YY41 pKa = 10.03WLDD44 pKa = 3.9NIQVYY49 pKa = 8.45PFRR52 pKa = 11.84IFIKK56 pKa = 10.52DD57 pKa = 3.62EE58 pKa = 3.7PHH60 pKa = 7.12KK61 pKa = 10.71LSKK64 pKa = 9.39KK65 pKa = 10.07QKK67 pKa = 9.34GRR69 pKa = 11.84WRR71 pKa = 11.84LIFSSPLFYY80 pKa = 10.73QILEE84 pKa = 4.22HH85 pKa = 7.18LLLDD89 pKa = 4.65PLDD92 pKa = 4.11EE93 pKa = 4.7MEE95 pKa = 6.42SLCQWDD101 pKa = 5.02LPTKK105 pKa = 10.38LRR107 pKa = 11.84WHH109 pKa = 6.74PFWGGAQMGVSNFDD123 pKa = 3.47NPVSLDD129 pKa = 3.57KK130 pKa = 11.31QCWDD134 pKa = 3.09WTLTGQFAEE143 pKa = 4.71LDD145 pKa = 3.35NALRR149 pKa = 11.84SNLVKK154 pKa = 10.74APNKK158 pKa = 7.54WHH160 pKa = 6.57EE161 pKa = 4.35LFEE164 pKa = 4.52TVNDD168 pKa = 4.88LIYY171 pKa = 11.12NKK173 pKa = 10.51AVFQFSSGHH182 pKa = 5.1QFQQVEE188 pKa = 4.25PGLMKK193 pKa = 10.61SGLVRR198 pKa = 11.84TLSTNSHH205 pKa = 5.59CQCFIHH211 pKa = 6.86WLAEE215 pKa = 4.19QKK217 pKa = 10.35CGHH220 pKa = 5.51SHH222 pKa = 6.45KK223 pKa = 10.26FWTIGDD229 pKa = 4.29DD230 pKa = 4.24LLVDD234 pKa = 4.88DD235 pKa = 5.52PCDD238 pKa = 3.84DD239 pKa = 3.84YY240 pKa = 11.97LRR242 pKa = 11.84YY243 pKa = 8.5TSDD246 pKa = 3.31YY247 pKa = 10.5CILKK251 pKa = 10.1EE252 pKa = 4.41VEE254 pKa = 3.84PGYY257 pKa = 11.27NFAGFNLKK265 pKa = 8.96TKK267 pKa = 9.96TPLYY271 pKa = 8.22WSKK274 pKa = 10.79HH275 pKa = 4.17IYY277 pKa = 9.45RR278 pKa = 11.84LLYY281 pKa = 10.06VDD283 pKa = 4.48EE284 pKa = 4.73KK285 pKa = 11.21VIPDD289 pKa = 3.59VLDD292 pKa = 3.62NYY294 pKa = 10.18QRR296 pKa = 11.84LYY298 pKa = 10.97VYY300 pKa = 10.61SDD302 pKa = 2.7KK303 pKa = 11.07KK304 pKa = 11.18FNIFQDD310 pKa = 3.44MLYY313 pKa = 10.79DD314 pKa = 4.68LDD316 pKa = 3.79PTKK319 pKa = 10.45IRR321 pKa = 11.84SQAYY325 pKa = 7.79LKK327 pKa = 10.23RR328 pKa = 11.84WAVQKK333 pKa = 10.44PSNFKK338 pKa = 10.66FLINN342 pKa = 3.65
MM1 pKa = 7.86DD2 pKa = 6.19ASPGLPFCKK11 pKa = 9.71HH12 pKa = 5.33YY13 pKa = 9.37KK14 pKa = 9.39TNAEE18 pKa = 4.09LLGFDD23 pKa = 5.18GFNCDD28 pKa = 4.91AVKK31 pKa = 10.64ADD33 pKa = 3.73CLVNMFYY40 pKa = 11.17YY41 pKa = 10.03WLDD44 pKa = 3.9NIQVYY49 pKa = 8.45PFRR52 pKa = 11.84IFIKK56 pKa = 10.52DD57 pKa = 3.62EE58 pKa = 3.7PHH60 pKa = 7.12KK61 pKa = 10.71LSKK64 pKa = 9.39KK65 pKa = 10.07QKK67 pKa = 9.34GRR69 pKa = 11.84WRR71 pKa = 11.84LIFSSPLFYY80 pKa = 10.73QILEE84 pKa = 4.22HH85 pKa = 7.18LLLDD89 pKa = 4.65PLDD92 pKa = 4.11EE93 pKa = 4.7MEE95 pKa = 6.42SLCQWDD101 pKa = 5.02LPTKK105 pKa = 10.38LRR107 pKa = 11.84WHH109 pKa = 6.74PFWGGAQMGVSNFDD123 pKa = 3.47NPVSLDD129 pKa = 3.57KK130 pKa = 11.31QCWDD134 pKa = 3.09WTLTGQFAEE143 pKa = 4.71LDD145 pKa = 3.35NALRR149 pKa = 11.84SNLVKK154 pKa = 10.74APNKK158 pKa = 7.54WHH160 pKa = 6.57EE161 pKa = 4.35LFEE164 pKa = 4.52TVNDD168 pKa = 4.88LIYY171 pKa = 11.12NKK173 pKa = 10.51AVFQFSSGHH182 pKa = 5.1QFQQVEE188 pKa = 4.25PGLMKK193 pKa = 10.61SGLVRR198 pKa = 11.84TLSTNSHH205 pKa = 5.59CQCFIHH211 pKa = 6.86WLAEE215 pKa = 4.19QKK217 pKa = 10.35CGHH220 pKa = 5.51SHH222 pKa = 6.45KK223 pKa = 10.26FWTIGDD229 pKa = 4.29DD230 pKa = 4.24LLVDD234 pKa = 4.88DD235 pKa = 5.52PCDD238 pKa = 3.84DD239 pKa = 3.84YY240 pKa = 11.97LRR242 pKa = 11.84YY243 pKa = 8.5TSDD246 pKa = 3.31YY247 pKa = 10.5CILKK251 pKa = 10.1EE252 pKa = 4.41VEE254 pKa = 3.84PGYY257 pKa = 11.27NFAGFNLKK265 pKa = 8.96TKK267 pKa = 9.96TPLYY271 pKa = 8.22WSKK274 pKa = 10.79HH275 pKa = 4.17IYY277 pKa = 9.45RR278 pKa = 11.84LLYY281 pKa = 10.06VDD283 pKa = 4.48EE284 pKa = 4.73KK285 pKa = 11.21VIPDD289 pKa = 3.59VLDD292 pKa = 3.62NYY294 pKa = 10.18QRR296 pKa = 11.84LYY298 pKa = 10.97VYY300 pKa = 10.61SDD302 pKa = 2.7KK303 pKa = 11.07KK304 pKa = 11.18FNIFQDD310 pKa = 3.44MLYY313 pKa = 10.79DD314 pKa = 4.68LDD316 pKa = 3.79PTKK319 pKa = 10.45IRR321 pKa = 11.84SQAYY325 pKa = 7.79LKK327 pKa = 10.23RR328 pKa = 11.84WAVQKK333 pKa = 10.44PSNFKK338 pKa = 10.66FLINN342 pKa = 3.65
Molecular weight: 40.42 kDa
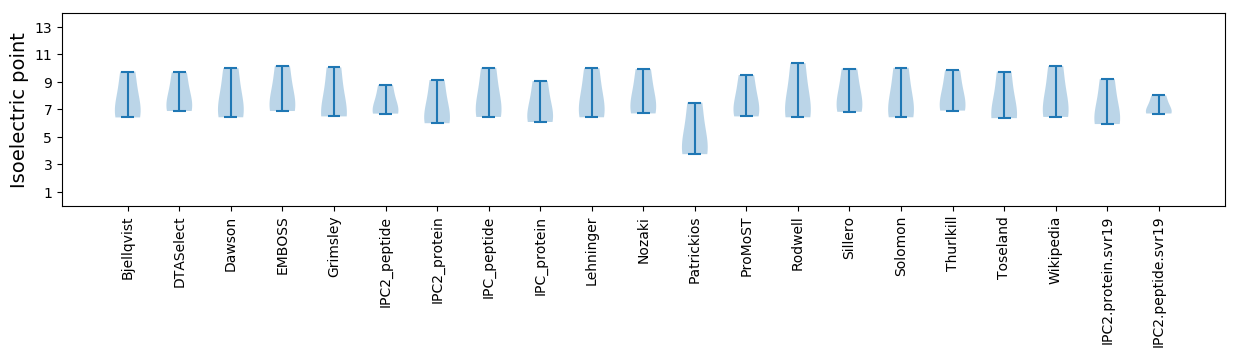
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEL5|A0A1L3KEL5_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 38 OX=1923225 PE=4 SV=1
MM1 pKa = 7.35TPLLPHH7 pKa = 6.75LQMNHH12 pKa = 4.47DD13 pKa = 4.73TIRR16 pKa = 11.84LEE18 pKa = 4.14SNLRR22 pKa = 11.84VYY24 pKa = 9.6GTVPNPSMQDD34 pKa = 3.29GPLHH38 pKa = 6.35NLYY41 pKa = 10.7CGLKK45 pKa = 10.14QKK47 pKa = 8.79TEE49 pKa = 4.13WNVMLALGGLEE60 pKa = 4.32TFWLLRR66 pKa = 11.84LIVYY70 pKa = 7.95LVKK73 pKa = 10.26MVGCVCLRR81 pKa = 11.84LLRR84 pKa = 11.84LFKK87 pKa = 10.55FVILRR92 pKa = 11.84LSIIMMNWQLWKK104 pKa = 11.1YY105 pKa = 10.9LFLCLLLYY113 pKa = 10.51KK114 pKa = 10.78LNLLKK119 pKa = 10.4FVPQMMQWFASQVVEE134 pKa = 6.01LYY136 pKa = 10.66DD137 pKa = 3.35QLPPVSSLLRR147 pKa = 11.84TFLEE151 pKa = 5.16CINILEE157 pKa = 4.31PQKK160 pKa = 11.09KK161 pKa = 9.94DD162 pKa = 3.17IPVLHH167 pKa = 6.89IWLEE171 pKa = 4.35TIQWPLCICSVVMLVICVCQPSTFKK196 pKa = 10.48WLSNNLLCQQSLGTSLGFMARR217 pKa = 11.84FWLRR221 pKa = 11.84QMTSLLAQASLNLSYY236 pKa = 10.03VTLVIVIPKK245 pKa = 10.01LIFVIKK251 pKa = 10.3DD252 pKa = 3.25YY253 pKa = 11.35LLNLVKK259 pKa = 10.64RR260 pKa = 11.84NANSRR265 pKa = 11.84SLMTGMKK272 pKa = 10.4KK273 pKa = 9.0MFSLAKK279 pKa = 10.46LFVLVVLGLTRR290 pKa = 11.84TVTNFNLAVIIILLTMMVFSGFGGLLISVVVRR322 pKa = 11.84LSWALLWALPLMVNPYY338 pKa = 9.56IIMMMFVLMKK348 pKa = 9.91MRR350 pKa = 11.84ILFF353 pKa = 3.98
MM1 pKa = 7.35TPLLPHH7 pKa = 6.75LQMNHH12 pKa = 4.47DD13 pKa = 4.73TIRR16 pKa = 11.84LEE18 pKa = 4.14SNLRR22 pKa = 11.84VYY24 pKa = 9.6GTVPNPSMQDD34 pKa = 3.29GPLHH38 pKa = 6.35NLYY41 pKa = 10.7CGLKK45 pKa = 10.14QKK47 pKa = 8.79TEE49 pKa = 4.13WNVMLALGGLEE60 pKa = 4.32TFWLLRR66 pKa = 11.84LIVYY70 pKa = 7.95LVKK73 pKa = 10.26MVGCVCLRR81 pKa = 11.84LLRR84 pKa = 11.84LFKK87 pKa = 10.55FVILRR92 pKa = 11.84LSIIMMNWQLWKK104 pKa = 11.1YY105 pKa = 10.9LFLCLLLYY113 pKa = 10.51KK114 pKa = 10.78LNLLKK119 pKa = 10.4FVPQMMQWFASQVVEE134 pKa = 6.01LYY136 pKa = 10.66DD137 pKa = 3.35QLPPVSSLLRR147 pKa = 11.84TFLEE151 pKa = 5.16CINILEE157 pKa = 4.31PQKK160 pKa = 11.09KK161 pKa = 9.94DD162 pKa = 3.17IPVLHH167 pKa = 6.89IWLEE171 pKa = 4.35TIQWPLCICSVVMLVICVCQPSTFKK196 pKa = 10.48WLSNNLLCQQSLGTSLGFMARR217 pKa = 11.84FWLRR221 pKa = 11.84QMTSLLAQASLNLSYY236 pKa = 10.03VTLVIVIPKK245 pKa = 10.01LIFVIKK251 pKa = 10.3DD252 pKa = 3.25YY253 pKa = 11.35LLNLVKK259 pKa = 10.64RR260 pKa = 11.84NANSRR265 pKa = 11.84SLMTGMKK272 pKa = 10.4KK273 pKa = 9.0MFSLAKK279 pKa = 10.46LFVLVVLGLTRR290 pKa = 11.84TVTNFNLAVIIILLTMMVFSGFGGLLISVVVRR322 pKa = 11.84LSWALLWALPLMVNPYY338 pKa = 9.56IIMMMFVLMKK348 pKa = 9.91MRR350 pKa = 11.84ILFF353 pKa = 3.98
Molecular weight: 40.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1181 |
342 |
486 |
393.7 |
45.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.234 ± 0.626 | 2.371 ± 0.32 |
5.419 ± 1.628 | 3.98 ± 0.815 |
5.165 ± 0.594 | 5.25 ± 0.788 |
1.778 ± 0.537 | 5.165 ± 0.571 |
6.097 ± 0.727 | 12.955 ± 2.976 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.556 ± 1.173 | 4.149 ± 0.567 |
4.742 ± 0.189 | 4.488 ± 0.182 |
4.657 ± 0.688 | 7.113 ± 0.946 |
5.165 ± 0.752 | 7.197 ± 1.014 |
2.625 ± 0.44 | 3.895 ± 0.629 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
