
Botrytis cinerea fusarivirus 1-S2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; Fusariviridae; Botrytis cinerea fusarivirus 1
Average proteome isoelectric point is 8.86
Get precalculated fractions of proteins
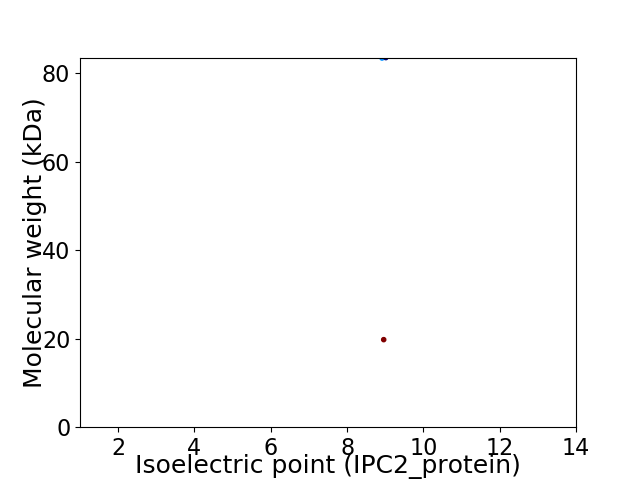
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U9NKQ1|A0A2U9NKQ1_9VIRU RdRp catalytic domain-containing protein (Fragment) OS=Botrytis cinerea fusarivirus 1-S2 OX=2219108 PE=4 SV=1
MM1 pKa = 7.75AFRR4 pKa = 11.84RR5 pKa = 11.84PWLRR9 pKa = 11.84LVLEE13 pKa = 4.53EE14 pKa = 3.99VTPPIEE20 pKa = 3.85DD21 pKa = 2.74RR22 pKa = 11.84MYY24 pKa = 10.97RR25 pKa = 11.84SVLALRR31 pKa = 11.84ALKK34 pKa = 9.84WLGAMGFMLPSDD46 pKa = 3.87SPTARR51 pKa = 11.84RR52 pKa = 11.84SRR54 pKa = 11.84IGTLSQLLQIGGKK67 pKa = 9.09IVNDD71 pKa = 3.54FDD73 pKa = 4.31LPAFIRR79 pKa = 11.84SPYY82 pKa = 9.31QSRR85 pKa = 11.84LTEE88 pKa = 4.01VEE90 pKa = 4.47TIDD93 pKa = 4.12LMKK96 pKa = 10.93SLGYY100 pKa = 7.18PTNAQVQIHH109 pKa = 5.8VPSKK113 pKa = 10.49SGQQNKK119 pKa = 8.66PWWIGRR125 pKa = 11.84SDD127 pKa = 3.72WKK129 pKa = 10.04MHH131 pKa = 6.09LPMFKK136 pKa = 10.58SFIQEE141 pKa = 3.74EE142 pKa = 4.53LEE144 pKa = 4.31DD145 pKa = 4.29FRR147 pKa = 11.84KK148 pKa = 9.97LRR150 pKa = 11.84NLPVFKK156 pKa = 10.52HH157 pKa = 5.98SYY159 pKa = 9.54TYY161 pKa = 9.77GTLDD165 pKa = 3.44SQVEE169 pKa = 4.47SISRR173 pKa = 11.84YY174 pKa = 8.65FYY176 pKa = 10.23EE177 pKa = 4.92SPVSMTLTKK186 pKa = 10.54DD187 pKa = 3.27DD188 pKa = 4.49YY189 pKa = 11.59DD190 pKa = 4.03IVWDD194 pKa = 4.07LVKK197 pKa = 10.63DD198 pKa = 3.87IYY200 pKa = 11.52ANSQLAPISEE210 pKa = 4.99IYY212 pKa = 10.37RR213 pKa = 11.84KK214 pKa = 8.61WDD216 pKa = 2.78KK217 pKa = 11.28KK218 pKa = 9.84MNVGVFASSRR228 pKa = 11.84EE229 pKa = 3.95KK230 pKa = 9.47FTKK233 pKa = 10.35FGSAKK238 pKa = 10.21KK239 pKa = 8.88MPRR242 pKa = 11.84WEE244 pKa = 4.55YY245 pKa = 8.89IRR247 pKa = 11.84LMGGHH252 pKa = 6.66KK253 pKa = 10.08KK254 pKa = 10.45VIKK257 pKa = 9.5QWYY260 pKa = 9.22AFFKK264 pKa = 10.27HH265 pKa = 5.44YY266 pKa = 10.77PRR268 pKa = 11.84MEE270 pKa = 3.89NFAQYY275 pKa = 7.87FTKK278 pKa = 10.87AEE280 pKa = 3.94FLPPKK285 pKa = 9.9KK286 pKa = 9.47WLNDD290 pKa = 3.45VVRR293 pKa = 11.84TPVASMLPQYY303 pKa = 10.02VSQMVWSGVQNHH315 pKa = 6.03RR316 pKa = 11.84FRR318 pKa = 11.84PFEE321 pKa = 4.1TPVKK325 pKa = 10.18IGLPINGTNLSKK337 pKa = 10.35IYY339 pKa = 10.35ALHH342 pKa = 6.43EE343 pKa = 4.13VTGGKK348 pKa = 10.05DD349 pKa = 3.28GATHH353 pKa = 7.16FAGDD357 pKa = 4.27CSAFDD362 pKa = 3.55STLTGPVLDD371 pKa = 4.55VIKK374 pKa = 10.37HH375 pKa = 3.84VRR377 pKa = 11.84KK378 pKa = 10.17RR379 pKa = 11.84GFKK382 pKa = 9.38DD383 pKa = 2.99HH384 pKa = 7.02RR385 pKa = 11.84AVKK388 pKa = 10.08AISWLVDD395 pKa = 3.24RR396 pKa = 11.84NYY398 pKa = 11.48AGIEE402 pKa = 3.78KK403 pKa = 10.42SLLVSSNTGDD413 pKa = 3.39IARR416 pKa = 11.84KK417 pKa = 9.91GSGLMTGHH425 pKa = 7.71ASTSADD431 pKa = 2.97NSLAMVALYY440 pKa = 10.92ALAWRR445 pKa = 11.84KK446 pKa = 7.96LTGTGSQEE454 pKa = 3.52FLKK457 pKa = 11.14YY458 pKa = 9.33NTLSVYY464 pKa = 10.86GDD466 pKa = 3.59DD467 pKa = 5.19HH468 pKa = 7.68ILSIARR474 pKa = 11.84NAPVQWTFPNVQSYY488 pKa = 9.62LASCNITLRR497 pKa = 11.84EE498 pKa = 3.85EE499 pKa = 4.23VQTGGKK505 pKa = 7.45GTKK508 pKa = 9.94LEE510 pKa = 4.66SIPFLSKK517 pKa = 10.01MARR520 pKa = 11.84KK521 pKa = 9.07PSAEE525 pKa = 3.93EE526 pKa = 3.36AHH528 pKa = 6.58LFEE531 pKa = 4.81SLGIEE536 pKa = 4.23VPRR539 pKa = 11.84WTISHH544 pKa = 6.93DD545 pKa = 3.56RR546 pKa = 11.84EE547 pKa = 4.06KK548 pKa = 11.27LLGKK552 pKa = 8.17MTAQLPNQNPLARR565 pKa = 11.84AKK567 pKa = 10.52RR568 pKa = 11.84MQSYY572 pKa = 10.56LYY574 pKa = 10.46LCAHH578 pKa = 6.18NPEE581 pKa = 4.64AYY583 pKa = 8.26EE584 pKa = 4.17ALTVALNTLFGKK596 pKa = 10.09FPKK599 pKa = 10.09VEE601 pKa = 4.16KK602 pKa = 10.43EE603 pKa = 3.8LGKK606 pKa = 9.01YY607 pKa = 6.86TPSYY611 pKa = 11.19ARR613 pKa = 11.84VLKK616 pKa = 10.29VWYY619 pKa = 9.69DD620 pKa = 3.47PEE622 pKa = 4.29SAPKK626 pKa = 9.45IDD628 pKa = 4.04EE629 pKa = 4.2DD630 pKa = 4.74HH631 pKa = 7.13LVGDD635 pKa = 4.72DD636 pKa = 4.98DD637 pKa = 4.95GVSAYY642 pKa = 10.45GGLTAMDD649 pKa = 3.53QFLNATAALPDD660 pKa = 3.86YY661 pKa = 9.94FNPIMKK667 pKa = 10.44NNGWVKK673 pKa = 10.55SIQQLAGTSLFWPNEE688 pKa = 3.86LIQRR692 pKa = 11.84TNSHH696 pKa = 5.89VSDD699 pKa = 3.97AQVSSSLRR707 pKa = 11.84KK708 pKa = 8.62SCYY711 pKa = 9.83SFLEE715 pKa = 3.94HH716 pKa = 7.14RR717 pKa = 11.84GTVPSSGSTLLTLLLRR733 pKa = 11.84HH734 pKa = 5.96WW735 pKa = 4.22
MM1 pKa = 7.75AFRR4 pKa = 11.84RR5 pKa = 11.84PWLRR9 pKa = 11.84LVLEE13 pKa = 4.53EE14 pKa = 3.99VTPPIEE20 pKa = 3.85DD21 pKa = 2.74RR22 pKa = 11.84MYY24 pKa = 10.97RR25 pKa = 11.84SVLALRR31 pKa = 11.84ALKK34 pKa = 9.84WLGAMGFMLPSDD46 pKa = 3.87SPTARR51 pKa = 11.84RR52 pKa = 11.84SRR54 pKa = 11.84IGTLSQLLQIGGKK67 pKa = 9.09IVNDD71 pKa = 3.54FDD73 pKa = 4.31LPAFIRR79 pKa = 11.84SPYY82 pKa = 9.31QSRR85 pKa = 11.84LTEE88 pKa = 4.01VEE90 pKa = 4.47TIDD93 pKa = 4.12LMKK96 pKa = 10.93SLGYY100 pKa = 7.18PTNAQVQIHH109 pKa = 5.8VPSKK113 pKa = 10.49SGQQNKK119 pKa = 8.66PWWIGRR125 pKa = 11.84SDD127 pKa = 3.72WKK129 pKa = 10.04MHH131 pKa = 6.09LPMFKK136 pKa = 10.58SFIQEE141 pKa = 3.74EE142 pKa = 4.53LEE144 pKa = 4.31DD145 pKa = 4.29FRR147 pKa = 11.84KK148 pKa = 9.97LRR150 pKa = 11.84NLPVFKK156 pKa = 10.52HH157 pKa = 5.98SYY159 pKa = 9.54TYY161 pKa = 9.77GTLDD165 pKa = 3.44SQVEE169 pKa = 4.47SISRR173 pKa = 11.84YY174 pKa = 8.65FYY176 pKa = 10.23EE177 pKa = 4.92SPVSMTLTKK186 pKa = 10.54DD187 pKa = 3.27DD188 pKa = 4.49YY189 pKa = 11.59DD190 pKa = 4.03IVWDD194 pKa = 4.07LVKK197 pKa = 10.63DD198 pKa = 3.87IYY200 pKa = 11.52ANSQLAPISEE210 pKa = 4.99IYY212 pKa = 10.37RR213 pKa = 11.84KK214 pKa = 8.61WDD216 pKa = 2.78KK217 pKa = 11.28KK218 pKa = 9.84MNVGVFASSRR228 pKa = 11.84EE229 pKa = 3.95KK230 pKa = 9.47FTKK233 pKa = 10.35FGSAKK238 pKa = 10.21KK239 pKa = 8.88MPRR242 pKa = 11.84WEE244 pKa = 4.55YY245 pKa = 8.89IRR247 pKa = 11.84LMGGHH252 pKa = 6.66KK253 pKa = 10.08KK254 pKa = 10.45VIKK257 pKa = 9.5QWYY260 pKa = 9.22AFFKK264 pKa = 10.27HH265 pKa = 5.44YY266 pKa = 10.77PRR268 pKa = 11.84MEE270 pKa = 3.89NFAQYY275 pKa = 7.87FTKK278 pKa = 10.87AEE280 pKa = 3.94FLPPKK285 pKa = 9.9KK286 pKa = 9.47WLNDD290 pKa = 3.45VVRR293 pKa = 11.84TPVASMLPQYY303 pKa = 10.02VSQMVWSGVQNHH315 pKa = 6.03RR316 pKa = 11.84FRR318 pKa = 11.84PFEE321 pKa = 4.1TPVKK325 pKa = 10.18IGLPINGTNLSKK337 pKa = 10.35IYY339 pKa = 10.35ALHH342 pKa = 6.43EE343 pKa = 4.13VTGGKK348 pKa = 10.05DD349 pKa = 3.28GATHH353 pKa = 7.16FAGDD357 pKa = 4.27CSAFDD362 pKa = 3.55STLTGPVLDD371 pKa = 4.55VIKK374 pKa = 10.37HH375 pKa = 3.84VRR377 pKa = 11.84KK378 pKa = 10.17RR379 pKa = 11.84GFKK382 pKa = 9.38DD383 pKa = 2.99HH384 pKa = 7.02RR385 pKa = 11.84AVKK388 pKa = 10.08AISWLVDD395 pKa = 3.24RR396 pKa = 11.84NYY398 pKa = 11.48AGIEE402 pKa = 3.78KK403 pKa = 10.42SLLVSSNTGDD413 pKa = 3.39IARR416 pKa = 11.84KK417 pKa = 9.91GSGLMTGHH425 pKa = 7.71ASTSADD431 pKa = 2.97NSLAMVALYY440 pKa = 10.92ALAWRR445 pKa = 11.84KK446 pKa = 7.96LTGTGSQEE454 pKa = 3.52FLKK457 pKa = 11.14YY458 pKa = 9.33NTLSVYY464 pKa = 10.86GDD466 pKa = 3.59DD467 pKa = 5.19HH468 pKa = 7.68ILSIARR474 pKa = 11.84NAPVQWTFPNVQSYY488 pKa = 9.62LASCNITLRR497 pKa = 11.84EE498 pKa = 3.85EE499 pKa = 4.23VQTGGKK505 pKa = 7.45GTKK508 pKa = 9.94LEE510 pKa = 4.66SIPFLSKK517 pKa = 10.01MARR520 pKa = 11.84KK521 pKa = 9.07PSAEE525 pKa = 3.93EE526 pKa = 3.36AHH528 pKa = 6.58LFEE531 pKa = 4.81SLGIEE536 pKa = 4.23VPRR539 pKa = 11.84WTISHH544 pKa = 6.93DD545 pKa = 3.56RR546 pKa = 11.84EE547 pKa = 4.06KK548 pKa = 11.27LLGKK552 pKa = 8.17MTAQLPNQNPLARR565 pKa = 11.84AKK567 pKa = 10.52RR568 pKa = 11.84MQSYY572 pKa = 10.56LYY574 pKa = 10.46LCAHH578 pKa = 6.18NPEE581 pKa = 4.64AYY583 pKa = 8.26EE584 pKa = 4.17ALTVALNTLFGKK596 pKa = 10.09FPKK599 pKa = 10.09VEE601 pKa = 4.16KK602 pKa = 10.43EE603 pKa = 3.8LGKK606 pKa = 9.01YY607 pKa = 6.86TPSYY611 pKa = 11.19ARR613 pKa = 11.84VLKK616 pKa = 10.29VWYY619 pKa = 9.69DD620 pKa = 3.47PEE622 pKa = 4.29SAPKK626 pKa = 9.45IDD628 pKa = 4.04EE629 pKa = 4.2DD630 pKa = 4.74HH631 pKa = 7.13LVGDD635 pKa = 4.72DD636 pKa = 4.98DD637 pKa = 4.95GVSAYY642 pKa = 10.45GGLTAMDD649 pKa = 3.53QFLNATAALPDD660 pKa = 3.86YY661 pKa = 9.94FNPIMKK667 pKa = 10.44NNGWVKK673 pKa = 10.55SIQQLAGTSLFWPNEE688 pKa = 3.86LIQRR692 pKa = 11.84TNSHH696 pKa = 5.89VSDD699 pKa = 3.97AQVSSSLRR707 pKa = 11.84KK708 pKa = 8.62SCYY711 pKa = 9.83SFLEE715 pKa = 3.94HH716 pKa = 7.14RR717 pKa = 11.84GTVPSSGSTLLTLLLRR733 pKa = 11.84HH734 pKa = 5.96WW735 pKa = 4.22
Molecular weight: 83.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U9NKQ1|A0A2U9NKQ1_9VIRU RdRp catalytic domain-containing protein (Fragment) OS=Botrytis cinerea fusarivirus 1-S2 OX=2219108 PE=4 SV=1
MM1 pKa = 7.35WSIIFSVYY9 pKa = 9.0WPEE12 pKa = 4.56AIQLIAATFWMWAAFIIVFPAMLILHH38 pKa = 6.73GLADD42 pKa = 3.64VMFICLFSLLVLRR55 pKa = 11.84SYY57 pKa = 10.77GASLIFIFFKK67 pKa = 10.61FSMWGTSPAFNIADD81 pKa = 3.56QLRR84 pKa = 11.84PIVTGLEE91 pKa = 3.43KK92 pKa = 10.21WLNIDD97 pKa = 4.68LMSHH101 pKa = 5.33YY102 pKa = 10.27EE103 pKa = 3.84RR104 pKa = 11.84AIRR107 pKa = 11.84DD108 pKa = 3.62YY109 pKa = 11.38SLSPWSLQFLIIFIISKK126 pKa = 9.84YY127 pKa = 8.25FLRR130 pKa = 11.84RR131 pKa = 11.84QMDD134 pKa = 3.92FAKK137 pKa = 10.55SLLWALLISNVLGPPLQAQGLLRR160 pKa = 11.84QSRR163 pKa = 11.84KK164 pKa = 9.36FCLGFII170 pKa = 4.39
MM1 pKa = 7.35WSIIFSVYY9 pKa = 9.0WPEE12 pKa = 4.56AIQLIAATFWMWAAFIIVFPAMLILHH38 pKa = 6.73GLADD42 pKa = 3.64VMFICLFSLLVLRR55 pKa = 11.84SYY57 pKa = 10.77GASLIFIFFKK67 pKa = 10.61FSMWGTSPAFNIADD81 pKa = 3.56QLRR84 pKa = 11.84PIVTGLEE91 pKa = 3.43KK92 pKa = 10.21WLNIDD97 pKa = 4.68LMSHH101 pKa = 5.33YY102 pKa = 10.27EE103 pKa = 3.84RR104 pKa = 11.84AIRR107 pKa = 11.84DD108 pKa = 3.62YY109 pKa = 11.38SLSPWSLQFLIIFIISKK126 pKa = 9.84YY127 pKa = 8.25FLRR130 pKa = 11.84RR131 pKa = 11.84QMDD134 pKa = 3.92FAKK137 pKa = 10.55SLLWALLISNVLGPPLQAQGLLRR160 pKa = 11.84QSRR163 pKa = 11.84KK164 pKa = 9.36FCLGFII170 pKa = 4.39
Molecular weight: 19.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
905 |
170 |
735 |
452.5 |
51.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.182 ± 0.497 | 0.663 ± 0.242 |
4.42 ± 0.698 | 4.199 ± 1.149 |
5.414 ± 2.165 | 5.746 ± 0.769 |
2.32 ± 0.54 | 5.856 ± 2.79 |
6.298 ± 1.585 | 11.05 ± 2.004 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.094 ± 0.483 | 3.425 ± 0.784 |
5.193 ± 0.508 | 3.757 ± 0.17 |
5.414 ± 0.335 | 8.619 ± 0.097 |
4.862 ± 1.462 | 5.635 ± 0.994 |
2.983 ± 0.813 | 3.867 ± 0.437 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
