
Human circovirus VS6600022
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
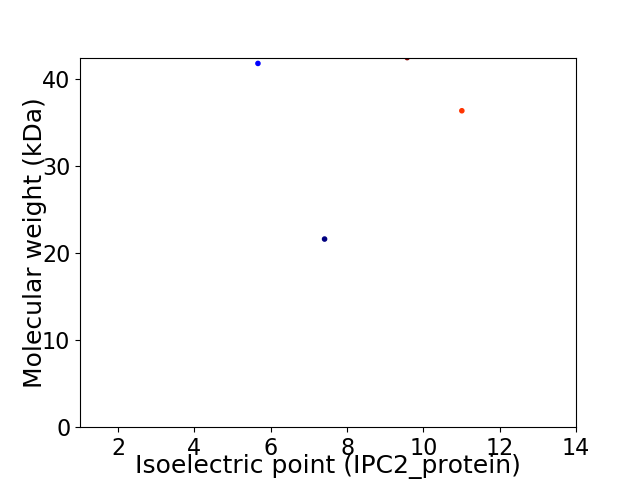
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0Q923|A0A0A0Q923_9CIRC Uncharacterized protein OS=Human circovirus VS6600022 OX=1525173 PE=4 SV=1
MM1 pKa = 7.54VYY3 pKa = 10.52NSSGKK8 pKa = 7.93YY9 pKa = 8.88RR10 pKa = 11.84KK11 pKa = 10.18VFVTSFNTSAMEE23 pKa = 4.06ALNIQDD29 pKa = 6.01LVDD32 pKa = 4.32CDD34 pKa = 3.54VARR37 pKa = 11.84YY38 pKa = 9.47VCMSMEE44 pKa = 4.37HH45 pKa = 6.7APTTGRR51 pKa = 11.84IHH53 pKa = 5.3YY54 pKa = 9.25HH55 pKa = 6.45LYY57 pKa = 10.23IEE59 pKa = 5.44FYY61 pKa = 9.73SQKK64 pKa = 9.59TMNTIKK70 pKa = 10.66KK71 pKa = 8.96ILKK74 pKa = 9.84DD75 pKa = 3.26ATANIQPARR84 pKa = 11.84GTPQEE89 pKa = 3.91AVDD92 pKa = 4.03YY93 pKa = 10.0IKK95 pKa = 10.92KK96 pKa = 10.65DD97 pKa = 3.0GATWKK102 pKa = 10.42EE103 pKa = 3.81AGTMSNQGHH112 pKa = 6.77RR113 pKa = 11.84SDD115 pKa = 5.3LDD117 pKa = 3.87DD118 pKa = 3.57VFQRR122 pKa = 11.84LQVGDD127 pKa = 4.46NILDD131 pKa = 4.09IIEE134 pKa = 4.3GHH136 pKa = 7.13PGTCIRR142 pKa = 11.84YY143 pKa = 8.83IKK145 pKa = 10.47GILAVKK151 pKa = 10.28GLVDD155 pKa = 3.89QRR157 pKa = 11.84RR158 pKa = 11.84QRR160 pKa = 11.84QEE162 pKa = 3.36ARR164 pKa = 11.84QMPTVLVYY172 pKa = 10.37IGKK175 pKa = 9.44SGAGKK180 pKa = 9.6SHH182 pKa = 7.1ACSQDD187 pKa = 2.76PDD189 pKa = 3.99YY190 pKa = 11.29QRR192 pKa = 11.84SGYY195 pKa = 9.38KK196 pKa = 10.22YY197 pKa = 9.54PVQGPSKK204 pKa = 10.69VYY206 pKa = 9.11FDD208 pKa = 4.88GYY210 pKa = 9.38TGEE213 pKa = 4.09STIWFDD219 pKa = 3.54EE220 pKa = 4.49FGGSVLPFHH229 pKa = 6.85VFLRR233 pKa = 11.84LADD236 pKa = 3.7KK237 pKa = 11.35YY238 pKa = 7.39EE239 pKa = 4.07TRR241 pKa = 11.84VEE243 pKa = 4.3TKK245 pKa = 10.31GGSVCITGLQKK256 pKa = 10.55ILISTTTPPKK266 pKa = 10.39LWWCEE271 pKa = 3.63SRR273 pKa = 11.84KK274 pKa = 9.97FNEE277 pKa = 4.98DD278 pKa = 3.31PYY280 pKa = 10.53QLWRR284 pKa = 11.84RR285 pKa = 11.84LTRR288 pKa = 11.84VYY290 pKa = 10.29YY291 pKa = 9.94IPRR294 pKa = 11.84PSWGFADD301 pKa = 3.87PVLIQHH307 pKa = 6.81PEE309 pKa = 3.83NFDD312 pKa = 3.62EE313 pKa = 5.71LVAAQLEE320 pKa = 4.71RR321 pKa = 11.84EE322 pKa = 4.34AEE324 pKa = 4.18TRR326 pKa = 11.84GPSLDD331 pKa = 3.68GEE333 pKa = 4.69SVRR336 pKa = 11.84GQPGPVDD343 pKa = 3.51PVHH346 pKa = 6.65EE347 pKa = 5.14RR348 pKa = 11.84DD349 pKa = 4.31DD350 pKa = 5.3DD351 pKa = 4.47SDD353 pKa = 4.06SEE355 pKa = 4.34RR356 pKa = 11.84TIDD359 pKa = 3.59TSDD362 pKa = 4.03GEE364 pKa = 4.54HH365 pKa = 6.23STTAAA370 pKa = 3.51
MM1 pKa = 7.54VYY3 pKa = 10.52NSSGKK8 pKa = 7.93YY9 pKa = 8.88RR10 pKa = 11.84KK11 pKa = 10.18VFVTSFNTSAMEE23 pKa = 4.06ALNIQDD29 pKa = 6.01LVDD32 pKa = 4.32CDD34 pKa = 3.54VARR37 pKa = 11.84YY38 pKa = 9.47VCMSMEE44 pKa = 4.37HH45 pKa = 6.7APTTGRR51 pKa = 11.84IHH53 pKa = 5.3YY54 pKa = 9.25HH55 pKa = 6.45LYY57 pKa = 10.23IEE59 pKa = 5.44FYY61 pKa = 9.73SQKK64 pKa = 9.59TMNTIKK70 pKa = 10.66KK71 pKa = 8.96ILKK74 pKa = 9.84DD75 pKa = 3.26ATANIQPARR84 pKa = 11.84GTPQEE89 pKa = 3.91AVDD92 pKa = 4.03YY93 pKa = 10.0IKK95 pKa = 10.92KK96 pKa = 10.65DD97 pKa = 3.0GATWKK102 pKa = 10.42EE103 pKa = 3.81AGTMSNQGHH112 pKa = 6.77RR113 pKa = 11.84SDD115 pKa = 5.3LDD117 pKa = 3.87DD118 pKa = 3.57VFQRR122 pKa = 11.84LQVGDD127 pKa = 4.46NILDD131 pKa = 4.09IIEE134 pKa = 4.3GHH136 pKa = 7.13PGTCIRR142 pKa = 11.84YY143 pKa = 8.83IKK145 pKa = 10.47GILAVKK151 pKa = 10.28GLVDD155 pKa = 3.89QRR157 pKa = 11.84RR158 pKa = 11.84QRR160 pKa = 11.84QEE162 pKa = 3.36ARR164 pKa = 11.84QMPTVLVYY172 pKa = 10.37IGKK175 pKa = 9.44SGAGKK180 pKa = 9.6SHH182 pKa = 7.1ACSQDD187 pKa = 2.76PDD189 pKa = 3.99YY190 pKa = 11.29QRR192 pKa = 11.84SGYY195 pKa = 9.38KK196 pKa = 10.22YY197 pKa = 9.54PVQGPSKK204 pKa = 10.69VYY206 pKa = 9.11FDD208 pKa = 4.88GYY210 pKa = 9.38TGEE213 pKa = 4.09STIWFDD219 pKa = 3.54EE220 pKa = 4.49FGGSVLPFHH229 pKa = 6.85VFLRR233 pKa = 11.84LADD236 pKa = 3.7KK237 pKa = 11.35YY238 pKa = 7.39EE239 pKa = 4.07TRR241 pKa = 11.84VEE243 pKa = 4.3TKK245 pKa = 10.31GGSVCITGLQKK256 pKa = 10.55ILISTTTPPKK266 pKa = 10.39LWWCEE271 pKa = 3.63SRR273 pKa = 11.84KK274 pKa = 9.97FNEE277 pKa = 4.98DD278 pKa = 3.31PYY280 pKa = 10.53QLWRR284 pKa = 11.84RR285 pKa = 11.84LTRR288 pKa = 11.84VYY290 pKa = 10.29YY291 pKa = 9.94IPRR294 pKa = 11.84PSWGFADD301 pKa = 3.87PVLIQHH307 pKa = 6.81PEE309 pKa = 3.83NFDD312 pKa = 3.62EE313 pKa = 5.71LVAAQLEE320 pKa = 4.71RR321 pKa = 11.84EE322 pKa = 4.34AEE324 pKa = 4.18TRR326 pKa = 11.84GPSLDD331 pKa = 3.68GEE333 pKa = 4.69SVRR336 pKa = 11.84GQPGPVDD343 pKa = 3.51PVHH346 pKa = 6.65EE347 pKa = 5.14RR348 pKa = 11.84DD349 pKa = 4.31DD350 pKa = 5.3DD351 pKa = 4.47SDD353 pKa = 4.06SEE355 pKa = 4.34RR356 pKa = 11.84TIDD359 pKa = 3.59TSDD362 pKa = 4.03GEE364 pKa = 4.54HH365 pKa = 6.23STTAAA370 pKa = 3.51
Molecular weight: 41.82 kDa
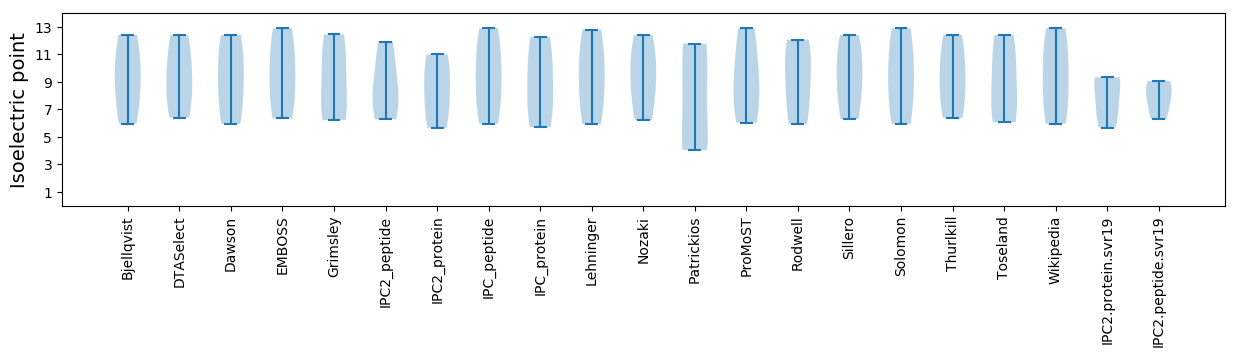
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0Q9Q5|A0A0A0Q9Q5_9CIRC Capsid protein OS=Human circovirus VS6600022 OX=1525173 PE=4 SV=1
MM1 pKa = 6.93VRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 9.62RR7 pKa = 11.84KK8 pKa = 8.01RR9 pKa = 11.84GRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VYY18 pKa = 10.41RR19 pKa = 11.84KK20 pKa = 9.33VRR22 pKa = 11.84LSKK25 pKa = 10.27SLYY28 pKa = 7.83RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.46IRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 7.67RR37 pKa = 11.84GKK39 pKa = 10.02RR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84GMTVSGTRR51 pKa = 11.84EE52 pKa = 4.0SQLVDD57 pKa = 3.38CSFIPLAKK65 pKa = 9.63PVLGTDD71 pKa = 2.86VGTPPGGFYY80 pKa = 9.53TWNINPLPTSVVNIPGDD97 pKa = 3.74PKK99 pKa = 10.93FVGAWSSDD107 pKa = 3.2PKK109 pKa = 11.0NILIRR114 pKa = 11.84QIGVGPNGTTEE125 pKa = 3.87TRR127 pKa = 11.84MLTDD131 pKa = 3.31MPVCISNVSVKK142 pKa = 10.86NIVRR146 pKa = 11.84CEE148 pKa = 4.23TMWKK152 pKa = 9.34LHH154 pKa = 5.4KK155 pKa = 10.14LYY157 pKa = 10.58RR158 pKa = 11.84ITFVSCTFTVPEE170 pKa = 4.01FTEE173 pKa = 4.57GKK175 pKa = 9.81RR176 pKa = 11.84NHH178 pKa = 6.19NLYY181 pKa = 10.82LEE183 pKa = 4.46WTHH186 pKa = 6.96LPRR189 pKa = 11.84GRR191 pKa = 11.84CAAYY195 pKa = 9.52EE196 pKa = 4.22DD197 pKa = 4.51CNGMVVGSAASSMNSFGFNWIVNPPDD223 pKa = 3.31IAEE226 pKa = 4.31ACCPDD231 pKa = 3.23GRR233 pKa = 11.84NNRR236 pKa = 11.84LNGWHH241 pKa = 6.72RR242 pKa = 11.84RR243 pKa = 11.84QLAYY247 pKa = 10.53NRR249 pKa = 11.84PVTISWRR256 pKa = 11.84PRR258 pKa = 11.84HH259 pKa = 5.14TNILEE264 pKa = 3.86DD265 pKa = 3.57HH266 pKa = 6.2QNYY269 pKa = 9.87LDD271 pKa = 4.48SIGSSGQGTMRR282 pKa = 11.84VLDD285 pKa = 3.65HH286 pKa = 7.11FGTKK290 pKa = 9.65NKK292 pKa = 9.33MIRR295 pKa = 11.84GYY297 pKa = 10.93LPTDD301 pKa = 3.05VDD303 pKa = 3.73QNIFDD308 pKa = 4.34EE309 pKa = 4.7RR310 pKa = 11.84QFWMGPVIRR319 pKa = 11.84LVDD322 pKa = 3.79ADD324 pKa = 4.69MPISDD329 pKa = 4.17QGGGINNKK337 pKa = 9.58SLFDD341 pKa = 3.66VYY343 pKa = 10.56GVRR346 pKa = 11.84VSTLVKK352 pKa = 10.85VKK354 pKa = 10.45FKK356 pKa = 11.41DD357 pKa = 3.48MNTTDD362 pKa = 4.07PLFPEE367 pKa = 4.61YY368 pKa = 11.07VPP370 pKa = 4.74
MM1 pKa = 6.93VRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 9.62RR7 pKa = 11.84KK8 pKa = 8.01RR9 pKa = 11.84GRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VYY18 pKa = 10.41RR19 pKa = 11.84KK20 pKa = 9.33VRR22 pKa = 11.84LSKK25 pKa = 10.27SLYY28 pKa = 7.83RR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.46IRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 7.67RR37 pKa = 11.84GKK39 pKa = 10.02RR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84GMTVSGTRR51 pKa = 11.84EE52 pKa = 4.0SQLVDD57 pKa = 3.38CSFIPLAKK65 pKa = 9.63PVLGTDD71 pKa = 2.86VGTPPGGFYY80 pKa = 9.53TWNINPLPTSVVNIPGDD97 pKa = 3.74PKK99 pKa = 10.93FVGAWSSDD107 pKa = 3.2PKK109 pKa = 11.0NILIRR114 pKa = 11.84QIGVGPNGTTEE125 pKa = 3.87TRR127 pKa = 11.84MLTDD131 pKa = 3.31MPVCISNVSVKK142 pKa = 10.86NIVRR146 pKa = 11.84CEE148 pKa = 4.23TMWKK152 pKa = 9.34LHH154 pKa = 5.4KK155 pKa = 10.14LYY157 pKa = 10.58RR158 pKa = 11.84ITFVSCTFTVPEE170 pKa = 4.01FTEE173 pKa = 4.57GKK175 pKa = 9.81RR176 pKa = 11.84NHH178 pKa = 6.19NLYY181 pKa = 10.82LEE183 pKa = 4.46WTHH186 pKa = 6.96LPRR189 pKa = 11.84GRR191 pKa = 11.84CAAYY195 pKa = 9.52EE196 pKa = 4.22DD197 pKa = 4.51CNGMVVGSAASSMNSFGFNWIVNPPDD223 pKa = 3.31IAEE226 pKa = 4.31ACCPDD231 pKa = 3.23GRR233 pKa = 11.84NNRR236 pKa = 11.84LNGWHH241 pKa = 6.72RR242 pKa = 11.84RR243 pKa = 11.84QLAYY247 pKa = 10.53NRR249 pKa = 11.84PVTISWRR256 pKa = 11.84PRR258 pKa = 11.84HH259 pKa = 5.14TNILEE264 pKa = 3.86DD265 pKa = 3.57HH266 pKa = 6.2QNYY269 pKa = 9.87LDD271 pKa = 4.48SIGSSGQGTMRR282 pKa = 11.84VLDD285 pKa = 3.65HH286 pKa = 7.11FGTKK290 pKa = 9.65NKK292 pKa = 9.33MIRR295 pKa = 11.84GYY297 pKa = 10.93LPTDD301 pKa = 3.05VDD303 pKa = 3.73QNIFDD308 pKa = 4.34EE309 pKa = 4.7RR310 pKa = 11.84QFWMGPVIRR319 pKa = 11.84LVDD322 pKa = 3.79ADD324 pKa = 4.69MPISDD329 pKa = 4.17QGGGINNKK337 pKa = 9.58SLFDD341 pKa = 3.66VYY343 pKa = 10.56GVRR346 pKa = 11.84VSTLVKK352 pKa = 10.85VKK354 pKa = 10.45FKK356 pKa = 11.41DD357 pKa = 3.48MNTTDD362 pKa = 4.07PLFPEE367 pKa = 4.61YY368 pKa = 11.07VPP370 pKa = 4.74
Molecular weight: 42.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1250 |
195 |
370 |
312.5 |
35.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.16 ± 0.657 | 1.52 ± 0.359 |
4.72 ± 1.384 | 3.36 ± 1.094 |
2.8 ± 0.43 | 6.08 ± 1.214 |
3.36 ± 0.785 | 7.2 ± 1.677 |
4.96 ± 0.276 | 6.56 ± 0.494 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.72 ± 0.514 | 4.72 ± 0.933 |
6.88 ± 0.858 | 4.16 ± 0.698 |
9.12 ± 1.518 | 8.4 ± 1.58 |
8.64 ± 1.707 | 5.6 ± 1.228 |
1.52 ± 0.359 | 3.52 ± 0.522 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
