
Albimonas donghaensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Albimonas
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
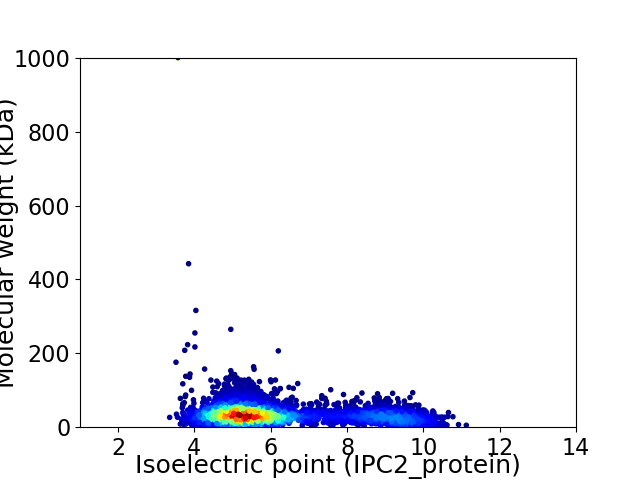
Virtual 2D-PAGE plot for 4253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3FFE6|A0A1H3FFE6_9RHOB Uncharacterized protein OS=Albimonas donghaensis OX=356660 GN=SAMN05444336_11287 PE=4 SV=1
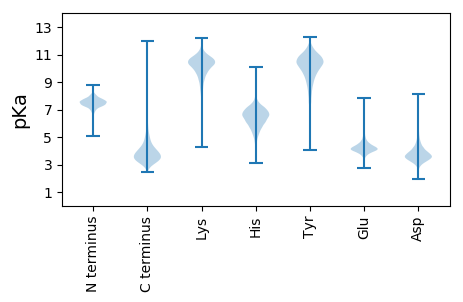
MM1 pKa = 6.95GTFAVTYY8 pKa = 9.65GYY10 pKa = 10.79FSFGGWSSSKK20 pKa = 10.7GGYY23 pKa = 9.22SSSGKK28 pKa = 9.99CDD30 pKa = 2.73WGYY33 pKa = 11.52SSSSWGGSSKK43 pKa = 10.75GSSHH47 pKa = 6.64GSKK50 pKa = 10.5GWGGHH55 pKa = 5.38NKK57 pKa = 10.2SNDD60 pKa = 3.51CDD62 pKa = 2.98WGYY65 pKa = 11.24GSKK68 pKa = 10.88GGGGGSKK75 pKa = 9.93KK76 pKa = 9.71HH77 pKa = 6.92DD78 pKa = 3.74SKK80 pKa = 11.45SWCDD84 pKa = 3.82DD85 pKa = 2.52GWGGSKK91 pKa = 10.64GGWGGSSWDD100 pKa = 3.45DD101 pKa = 3.76CGWGGGHH108 pKa = 6.55SGGGHH113 pKa = 6.22GGGHH117 pKa = 6.7GGGGHH122 pKa = 6.81GGGGHH127 pKa = 6.15GGGWGGGGWGGGWGCDD143 pKa = 3.42VNAEE147 pKa = 4.26PVFEE151 pKa = 5.39DD152 pKa = 4.2DD153 pKa = 4.73CNACIEE159 pKa = 4.19VTIDD163 pKa = 3.24EE164 pKa = 4.4NTLAVIDD171 pKa = 5.1LDD173 pKa = 3.87ATDD176 pKa = 4.92ADD178 pKa = 4.68GDD180 pKa = 4.05TLTYY184 pKa = 9.9EE185 pKa = 4.07ISGGDD190 pKa = 3.57DD191 pKa = 2.99AALFTVDD198 pKa = 3.94PATGEE203 pKa = 4.09LTFIDD208 pKa = 4.63APDD211 pKa = 4.06FEE213 pKa = 6.46APLDD217 pKa = 4.03ADD219 pKa = 3.62GDD221 pKa = 3.84NVYY224 pKa = 10.64DD225 pKa = 3.87VKK227 pKa = 11.6VKK229 pKa = 9.67VTDD232 pKa = 3.9GEE234 pKa = 4.51GGSDD238 pKa = 3.56EE239 pKa = 5.22ACVKK243 pKa = 10.79VEE245 pKa = 4.37VEE247 pKa = 4.95DD248 pKa = 5.04VDD250 pKa = 4.24DD251 pKa = 4.48TPPPSGGGACQFTLSQDD268 pKa = 3.39GLDD271 pKa = 3.81IEE273 pKa = 4.64VHH275 pKa = 5.07LTEE278 pKa = 5.48LPDD281 pKa = 3.85GSVQIEE287 pKa = 4.25LAVADD292 pKa = 4.96DD293 pKa = 3.89SAKK296 pKa = 11.02VGDD299 pKa = 3.68LRR301 pKa = 11.84GLFFHH306 pKa = 6.78VADD309 pKa = 4.11EE310 pKa = 4.45SKK312 pKa = 10.56IGGLRR317 pKa = 11.84ITGDD321 pKa = 3.79DD322 pKa = 3.52VTGTQYY328 pKa = 11.29CEE330 pKa = 4.97DD331 pKa = 3.8RR332 pKa = 11.84VKK334 pKa = 11.26DD335 pKa = 4.09LGWGANVSGLGRR347 pKa = 11.84FDD349 pKa = 4.81AGVKK353 pKa = 10.3LGTSGIGKK361 pKa = 9.83DD362 pKa = 4.23DD363 pKa = 3.4IRR365 pKa = 11.84STTLVISHH373 pKa = 7.88PDD375 pKa = 3.28GMSLDD380 pKa = 4.03DD381 pKa = 5.77FRR383 pKa = 11.84NQDD386 pKa = 3.08FAIRR390 pKa = 11.84ATSVGYY396 pKa = 10.13EE397 pKa = 4.15GGWRR401 pKa = 11.84GDD403 pKa = 3.53SEE405 pKa = 5.24KK406 pKa = 10.7IGGNAGEE413 pKa = 4.66LDD415 pKa = 3.85CGGANANSAPIAVDD429 pKa = 4.35DD430 pKa = 4.51EE431 pKa = 5.14GGLCSTDD438 pKa = 2.77ATLIDD443 pKa = 3.94VLANDD448 pKa = 4.19SDD450 pKa = 4.27ADD452 pKa = 3.91PDD454 pKa = 4.25EE455 pKa = 4.26VWGLLGIKK463 pKa = 8.84TDD465 pKa = 3.49AGIEE469 pKa = 4.11FMTDD473 pKa = 2.66GDD475 pKa = 4.97SFTLDD480 pKa = 2.66SGAVVTRR487 pKa = 11.84VGDD490 pKa = 3.45AFEE493 pKa = 4.81YY494 pKa = 10.73DD495 pKa = 3.43LSGTGDD501 pKa = 3.92DD502 pKa = 5.45FDD504 pKa = 4.52TLLVGQKK511 pKa = 10.39GADD514 pKa = 3.21SFEE517 pKa = 4.2YY518 pKa = 10.91VIGDD522 pKa = 3.89DD523 pKa = 5.01EE524 pKa = 4.71GATDD528 pKa = 3.3IGEE531 pKa = 4.22VVIDD535 pKa = 3.25ICGGVNAVEE544 pKa = 6.1LIDD547 pKa = 4.6NDD549 pKa = 3.98NPLLASMDD557 pKa = 3.93LGQVLDD563 pKa = 3.91PVSWLVEE570 pKa = 4.22TVNAAVDD577 pKa = 4.13FDD579 pKa = 3.93EE580 pKa = 4.79TAAEE584 pKa = 4.05ALLDD588 pKa = 4.24LGALGLTDD596 pKa = 4.25HH597 pKa = 7.2GIDD600 pKa = 3.64GLFGPAYY607 pKa = 10.06CVDD610 pKa = 3.37RR611 pKa = 11.84TRR613 pKa = 11.84PANYY617 pKa = 9.15DD618 pKa = 3.14ATDD621 pKa = 3.45ALVYY625 pKa = 10.08TSTGDD630 pKa = 3.32IPDD633 pKa = 3.56VNFDD637 pKa = 3.7GGDD640 pKa = 3.67VIDD643 pKa = 5.39KK644 pKa = 10.83EE645 pKa = 4.46EE646 pKa = 4.65NLDD649 pKa = 3.7NLNWLLSQRR658 pKa = 11.84YY659 pKa = 7.91EE660 pKa = 4.13DD661 pKa = 3.31QGYY664 pKa = 9.7GYY666 pKa = 9.07TDD668 pKa = 3.11IQSAIWQLMDD678 pKa = 4.11DD679 pKa = 3.99VGGIDD684 pKa = 3.73TSRR687 pKa = 11.84STPGVQLTAAAQTMYY702 pKa = 10.65DD703 pKa = 3.1AAMANGDD710 pKa = 3.63GFAPTGADD718 pKa = 3.19GDD720 pKa = 4.16VLGLIFQPVSDD731 pKa = 4.54NGSGGYY737 pKa = 9.24DD738 pKa = 3.02ANGQIFVVGYY748 pKa = 10.19DD749 pKa = 3.91LQDD752 pKa = 3.79CLCYY756 pKa = 10.58
MM1 pKa = 6.95GTFAVTYY8 pKa = 9.65GYY10 pKa = 10.79FSFGGWSSSKK20 pKa = 10.7GGYY23 pKa = 9.22SSSGKK28 pKa = 9.99CDD30 pKa = 2.73WGYY33 pKa = 11.52SSSSWGGSSKK43 pKa = 10.75GSSHH47 pKa = 6.64GSKK50 pKa = 10.5GWGGHH55 pKa = 5.38NKK57 pKa = 10.2SNDD60 pKa = 3.51CDD62 pKa = 2.98WGYY65 pKa = 11.24GSKK68 pKa = 10.88GGGGGSKK75 pKa = 9.93KK76 pKa = 9.71HH77 pKa = 6.92DD78 pKa = 3.74SKK80 pKa = 11.45SWCDD84 pKa = 3.82DD85 pKa = 2.52GWGGSKK91 pKa = 10.64GGWGGSSWDD100 pKa = 3.45DD101 pKa = 3.76CGWGGGHH108 pKa = 6.55SGGGHH113 pKa = 6.22GGGHH117 pKa = 6.7GGGGHH122 pKa = 6.81GGGGHH127 pKa = 6.15GGGWGGGGWGGGWGCDD143 pKa = 3.42VNAEE147 pKa = 4.26PVFEE151 pKa = 5.39DD152 pKa = 4.2DD153 pKa = 4.73CNACIEE159 pKa = 4.19VTIDD163 pKa = 3.24EE164 pKa = 4.4NTLAVIDD171 pKa = 5.1LDD173 pKa = 3.87ATDD176 pKa = 4.92ADD178 pKa = 4.68GDD180 pKa = 4.05TLTYY184 pKa = 9.9EE185 pKa = 4.07ISGGDD190 pKa = 3.57DD191 pKa = 2.99AALFTVDD198 pKa = 3.94PATGEE203 pKa = 4.09LTFIDD208 pKa = 4.63APDD211 pKa = 4.06FEE213 pKa = 6.46APLDD217 pKa = 4.03ADD219 pKa = 3.62GDD221 pKa = 3.84NVYY224 pKa = 10.64DD225 pKa = 3.87VKK227 pKa = 11.6VKK229 pKa = 9.67VTDD232 pKa = 3.9GEE234 pKa = 4.51GGSDD238 pKa = 3.56EE239 pKa = 5.22ACVKK243 pKa = 10.79VEE245 pKa = 4.37VEE247 pKa = 4.95DD248 pKa = 5.04VDD250 pKa = 4.24DD251 pKa = 4.48TPPPSGGGACQFTLSQDD268 pKa = 3.39GLDD271 pKa = 3.81IEE273 pKa = 4.64VHH275 pKa = 5.07LTEE278 pKa = 5.48LPDD281 pKa = 3.85GSVQIEE287 pKa = 4.25LAVADD292 pKa = 4.96DD293 pKa = 3.89SAKK296 pKa = 11.02VGDD299 pKa = 3.68LRR301 pKa = 11.84GLFFHH306 pKa = 6.78VADD309 pKa = 4.11EE310 pKa = 4.45SKK312 pKa = 10.56IGGLRR317 pKa = 11.84ITGDD321 pKa = 3.79DD322 pKa = 3.52VTGTQYY328 pKa = 11.29CEE330 pKa = 4.97DD331 pKa = 3.8RR332 pKa = 11.84VKK334 pKa = 11.26DD335 pKa = 4.09LGWGANVSGLGRR347 pKa = 11.84FDD349 pKa = 4.81AGVKK353 pKa = 10.3LGTSGIGKK361 pKa = 9.83DD362 pKa = 4.23DD363 pKa = 3.4IRR365 pKa = 11.84STTLVISHH373 pKa = 7.88PDD375 pKa = 3.28GMSLDD380 pKa = 4.03DD381 pKa = 5.77FRR383 pKa = 11.84NQDD386 pKa = 3.08FAIRR390 pKa = 11.84ATSVGYY396 pKa = 10.13EE397 pKa = 4.15GGWRR401 pKa = 11.84GDD403 pKa = 3.53SEE405 pKa = 5.24KK406 pKa = 10.7IGGNAGEE413 pKa = 4.66LDD415 pKa = 3.85CGGANANSAPIAVDD429 pKa = 4.35DD430 pKa = 4.51EE431 pKa = 5.14GGLCSTDD438 pKa = 2.77ATLIDD443 pKa = 3.94VLANDD448 pKa = 4.19SDD450 pKa = 4.27ADD452 pKa = 3.91PDD454 pKa = 4.25EE455 pKa = 4.26VWGLLGIKK463 pKa = 8.84TDD465 pKa = 3.49AGIEE469 pKa = 4.11FMTDD473 pKa = 2.66GDD475 pKa = 4.97SFTLDD480 pKa = 2.66SGAVVTRR487 pKa = 11.84VGDD490 pKa = 3.45AFEE493 pKa = 4.81YY494 pKa = 10.73DD495 pKa = 3.43LSGTGDD501 pKa = 3.92DD502 pKa = 5.45FDD504 pKa = 4.52TLLVGQKK511 pKa = 10.39GADD514 pKa = 3.21SFEE517 pKa = 4.2YY518 pKa = 10.91VIGDD522 pKa = 3.89DD523 pKa = 5.01EE524 pKa = 4.71GATDD528 pKa = 3.3IGEE531 pKa = 4.22VVIDD535 pKa = 3.25ICGGVNAVEE544 pKa = 6.1LIDD547 pKa = 4.6NDD549 pKa = 3.98NPLLASMDD557 pKa = 3.93LGQVLDD563 pKa = 3.91PVSWLVEE570 pKa = 4.22TVNAAVDD577 pKa = 4.13FDD579 pKa = 3.93EE580 pKa = 4.79TAAEE584 pKa = 4.05ALLDD588 pKa = 4.24LGALGLTDD596 pKa = 4.25HH597 pKa = 7.2GIDD600 pKa = 3.64GLFGPAYY607 pKa = 10.06CVDD610 pKa = 3.37RR611 pKa = 11.84TRR613 pKa = 11.84PANYY617 pKa = 9.15DD618 pKa = 3.14ATDD621 pKa = 3.45ALVYY625 pKa = 10.08TSTGDD630 pKa = 3.32IPDD633 pKa = 3.56VNFDD637 pKa = 3.7GGDD640 pKa = 3.67VIDD643 pKa = 5.39KK644 pKa = 10.83EE645 pKa = 4.46EE646 pKa = 4.65NLDD649 pKa = 3.7NLNWLLSQRR658 pKa = 11.84YY659 pKa = 7.91EE660 pKa = 4.13DD661 pKa = 3.31QGYY664 pKa = 9.7GYY666 pKa = 9.07TDD668 pKa = 3.11IQSAIWQLMDD678 pKa = 4.11DD679 pKa = 3.99VGGIDD684 pKa = 3.73TSRR687 pKa = 11.84STPGVQLTAAAQTMYY702 pKa = 10.65DD703 pKa = 3.1AAMANGDD710 pKa = 3.63GFAPTGADD718 pKa = 3.19GDD720 pKa = 4.16VLGLIFQPVSDD731 pKa = 4.54NGSGGYY737 pKa = 9.24DD738 pKa = 3.02ANGQIFVVGYY748 pKa = 10.19DD749 pKa = 3.91LQDD752 pKa = 3.79CLCYY756 pKa = 10.58
Molecular weight: 77.94 kDa
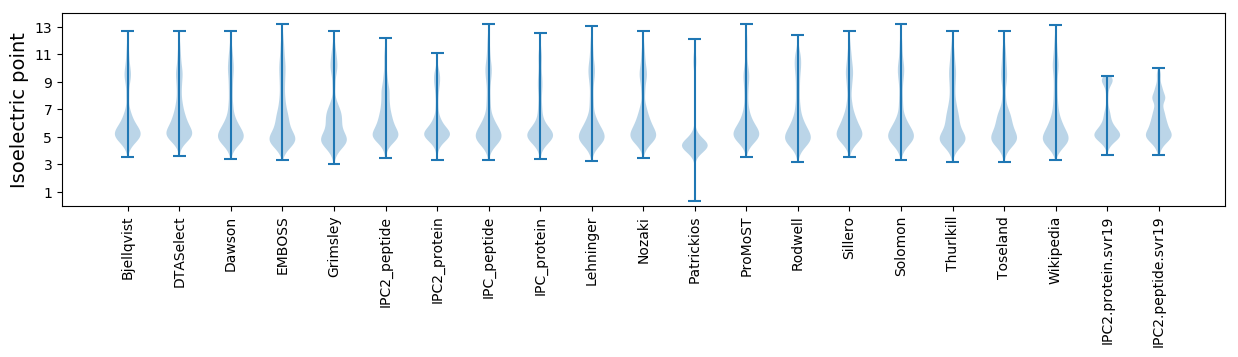
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3A3J3|A0A1H3A3J3_9RHOB Histidine kinase OS=Albimonas donghaensis OX=356660 GN=SAMN05444336_10423 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42NGAKK29 pKa = 9.88IIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.25GRR39 pKa = 11.84HH40 pKa = 5.49KK41 pKa = 10.9LSAA44 pKa = 3.8
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42NGAKK29 pKa = 9.88IIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.25GRR39 pKa = 11.84HH40 pKa = 5.49KK41 pKa = 10.9LSAA44 pKa = 3.8
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1441178 |
31 |
9789 |
338.9 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.395 ± 0.082 | 0.817 ± 0.013 |
5.96 ± 0.052 | 5.999 ± 0.038 |
3.442 ± 0.019 | 9.762 ± 0.078 |
1.799 ± 0.018 | 4.259 ± 0.028 |
2.251 ± 0.036 | 9.971 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.576 ± 0.022 | 1.931 ± 0.023 |
5.964 ± 0.049 | 2.349 ± 0.022 |
7.777 ± 0.058 | 4.722 ± 0.035 |
4.819 ± 0.033 | 6.951 ± 0.035 |
1.424 ± 0.018 | 1.83 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
