
Human fecal virus Jorvi4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
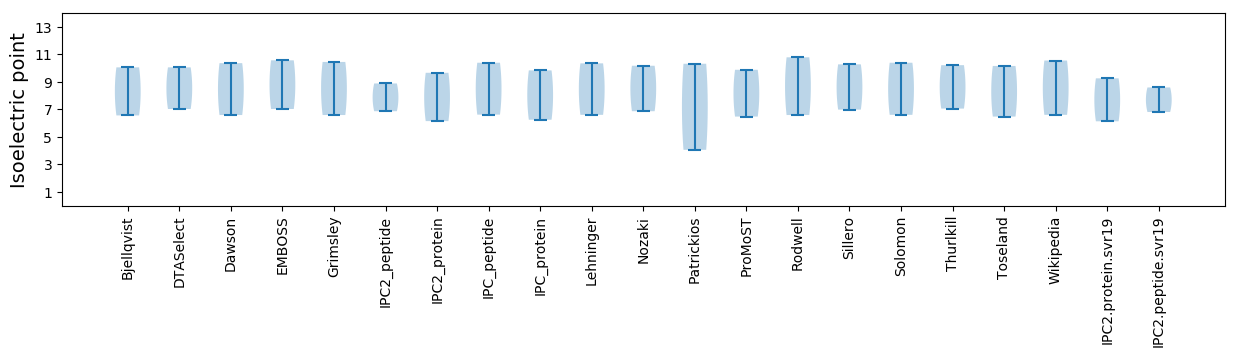
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A220IGD7|A0A220IGD7_9CIRC Uncharacterized protein OS=Human fecal virus Jorvi4 OX=2017083 PE=4 SV=1
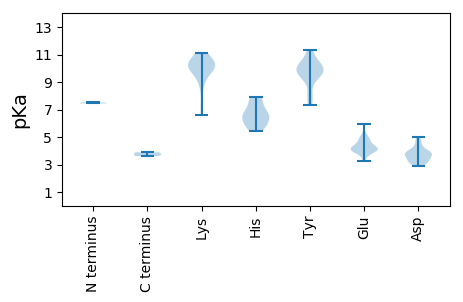
MM1 pKa = 7.44SRR3 pKa = 11.84YY4 pKa = 9.7QSICFTIFDD13 pKa = 3.92PHH15 pKa = 7.66IVNDD19 pKa = 4.56DD20 pKa = 3.7QVCMQRR26 pKa = 11.84AYY28 pKa = 10.02DD29 pKa = 3.63TGKK32 pKa = 10.82LGFLVYY38 pKa = 10.47QIEE41 pKa = 4.36RR42 pKa = 11.84CPNTNRR48 pKa = 11.84EE49 pKa = 4.33HH50 pKa = 5.58IQGYY54 pKa = 10.41AEE56 pKa = 3.99LHH58 pKa = 5.95SRR60 pKa = 11.84TTLGRR65 pKa = 11.84HH66 pKa = 5.65GPRR69 pKa = 11.84SSGVKK74 pKa = 10.33GIFRR78 pKa = 11.84SDD80 pKa = 3.12TMHH83 pKa = 7.9VEE85 pKa = 4.05MAKK88 pKa = 10.68GSAEE92 pKa = 4.01QNITYY97 pKa = 7.63CTKK100 pKa = 8.95PEE102 pKa = 3.86SRR104 pKa = 11.84VRR106 pKa = 11.84EE107 pKa = 4.11GLRR110 pKa = 11.84LGEE113 pKa = 4.3PANKK117 pKa = 10.09RR118 pKa = 11.84PGEE121 pKa = 4.09RR122 pKa = 11.84KK123 pKa = 9.7DD124 pKa = 3.25WEE126 pKa = 4.31EE127 pKa = 4.15LYY129 pKa = 10.1EE130 pKa = 4.16HH131 pKa = 7.42CIHH134 pKa = 7.81ADD136 pKa = 3.26TDD138 pKa = 3.9EE139 pKa = 4.71KK140 pKa = 11.04NLCIGHH146 pKa = 6.79PVLIKK151 pKa = 9.73YY152 pKa = 8.46YY153 pKa = 10.53RR154 pKa = 11.84SATEE158 pKa = 3.29IWSRR162 pKa = 11.84THH164 pKa = 6.28PPSMRR169 pKa = 11.84FPIRR173 pKa = 11.84IFILVGPTRR182 pKa = 11.84TGKK185 pKa = 8.0TRR187 pKa = 11.84WAFTKK192 pKa = 10.47FPEE195 pKa = 4.84LYY197 pKa = 10.13RR198 pKa = 11.84LCTEE202 pKa = 5.1KK203 pKa = 11.05ADD205 pKa = 3.16WWNGYY210 pKa = 9.35LGQKK214 pKa = 10.03DD215 pKa = 3.81VLIDD219 pKa = 3.64EE220 pKa = 5.17FDD222 pKa = 3.95GEE224 pKa = 4.87SIKK227 pKa = 9.96LTYY230 pKa = 9.57MLQLLDD236 pKa = 4.68RR237 pKa = 11.84YY238 pKa = 9.84PMQVPTKK245 pKa = 9.37GGFVRR250 pKa = 11.84LCATNWIITSNIDD263 pKa = 2.92HH264 pKa = 6.74DD265 pKa = 3.92QWYY268 pKa = 11.27SMAPQGNRR276 pKa = 11.84DD277 pKa = 3.17AFYY280 pKa = 11.36ARR282 pKa = 11.84IQEE285 pKa = 4.09WGRR288 pKa = 11.84VLSVVEE294 pKa = 4.0YY295 pKa = 10.97RR296 pKa = 11.84EE297 pKa = 4.15DD298 pKa = 3.47LQLEE302 pKa = 4.31EE303 pKa = 4.56QEE305 pKa = 4.46GAQAQEE311 pKa = 3.89
MM1 pKa = 7.44SRR3 pKa = 11.84YY4 pKa = 9.7QSICFTIFDD13 pKa = 3.92PHH15 pKa = 7.66IVNDD19 pKa = 4.56DD20 pKa = 3.7QVCMQRR26 pKa = 11.84AYY28 pKa = 10.02DD29 pKa = 3.63TGKK32 pKa = 10.82LGFLVYY38 pKa = 10.47QIEE41 pKa = 4.36RR42 pKa = 11.84CPNTNRR48 pKa = 11.84EE49 pKa = 4.33HH50 pKa = 5.58IQGYY54 pKa = 10.41AEE56 pKa = 3.99LHH58 pKa = 5.95SRR60 pKa = 11.84TTLGRR65 pKa = 11.84HH66 pKa = 5.65GPRR69 pKa = 11.84SSGVKK74 pKa = 10.33GIFRR78 pKa = 11.84SDD80 pKa = 3.12TMHH83 pKa = 7.9VEE85 pKa = 4.05MAKK88 pKa = 10.68GSAEE92 pKa = 4.01QNITYY97 pKa = 7.63CTKK100 pKa = 8.95PEE102 pKa = 3.86SRR104 pKa = 11.84VRR106 pKa = 11.84EE107 pKa = 4.11GLRR110 pKa = 11.84LGEE113 pKa = 4.3PANKK117 pKa = 10.09RR118 pKa = 11.84PGEE121 pKa = 4.09RR122 pKa = 11.84KK123 pKa = 9.7DD124 pKa = 3.25WEE126 pKa = 4.31EE127 pKa = 4.15LYY129 pKa = 10.1EE130 pKa = 4.16HH131 pKa = 7.42CIHH134 pKa = 7.81ADD136 pKa = 3.26TDD138 pKa = 3.9EE139 pKa = 4.71KK140 pKa = 11.04NLCIGHH146 pKa = 6.79PVLIKK151 pKa = 9.73YY152 pKa = 8.46YY153 pKa = 10.53RR154 pKa = 11.84SATEE158 pKa = 3.29IWSRR162 pKa = 11.84THH164 pKa = 6.28PPSMRR169 pKa = 11.84FPIRR173 pKa = 11.84IFILVGPTRR182 pKa = 11.84TGKK185 pKa = 8.0TRR187 pKa = 11.84WAFTKK192 pKa = 10.47FPEE195 pKa = 4.84LYY197 pKa = 10.13RR198 pKa = 11.84LCTEE202 pKa = 5.1KK203 pKa = 11.05ADD205 pKa = 3.16WWNGYY210 pKa = 9.35LGQKK214 pKa = 10.03DD215 pKa = 3.81VLIDD219 pKa = 3.64EE220 pKa = 5.17FDD222 pKa = 3.95GEE224 pKa = 4.87SIKK227 pKa = 9.96LTYY230 pKa = 9.57MLQLLDD236 pKa = 4.68RR237 pKa = 11.84YY238 pKa = 9.84PMQVPTKK245 pKa = 9.37GGFVRR250 pKa = 11.84LCATNWIITSNIDD263 pKa = 2.92HH264 pKa = 6.74DD265 pKa = 3.92QWYY268 pKa = 11.27SMAPQGNRR276 pKa = 11.84DD277 pKa = 3.17AFYY280 pKa = 11.36ARR282 pKa = 11.84IQEE285 pKa = 4.09WGRR288 pKa = 11.84VLSVVEE294 pKa = 4.0YY295 pKa = 10.97RR296 pKa = 11.84EE297 pKa = 4.15DD298 pKa = 3.47LQLEE302 pKa = 4.31EE303 pKa = 4.56QEE305 pKa = 4.46GAQAQEE311 pKa = 3.89
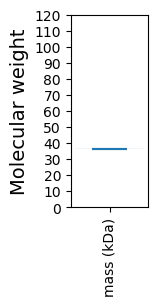
Molecular weight: 36.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220IGD7|A0A220IGD7_9CIRC Uncharacterized protein OS=Human fecal virus Jorvi4 OX=2017083 PE=4 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84RR4 pKa = 11.84GYY6 pKa = 10.1RR7 pKa = 11.84YY8 pKa = 9.73KK9 pKa = 10.53RR10 pKa = 11.84YY11 pKa = 9.38YY12 pKa = 9.85GKK14 pKa = 9.86NRR16 pKa = 11.84KK17 pKa = 9.07RR18 pKa = 11.84FIGPLLPGGYY28 pKa = 10.32AKK30 pKa = 10.93GNAKK34 pKa = 10.07SLARR38 pKa = 11.84KK39 pKa = 6.58WRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84YY45 pKa = 9.89RR46 pKa = 11.84YY47 pKa = 9.48GKK49 pKa = 10.01KK50 pKa = 9.92SKK52 pKa = 10.37QSLRR56 pKa = 11.84SGWGLGMTRR65 pKa = 11.84AKK67 pKa = 10.75SFVKK71 pKa = 10.32RR72 pKa = 11.84VDD74 pKa = 3.86LNEE77 pKa = 3.93FTPKK81 pKa = 9.98GSCRR85 pKa = 11.84FRR87 pKa = 11.84ISYY90 pKa = 10.1RR91 pKa = 11.84GRR93 pKa = 11.84HH94 pKa = 6.51LITAANVYY102 pKa = 9.68CAANFHH108 pKa = 6.22PHH110 pKa = 5.4PHH112 pKa = 5.94YY113 pKa = 8.05PTTGFADD120 pKa = 3.86PVLSATSDD128 pKa = 3.58DD129 pKa = 3.98FTSDD133 pKa = 3.35PRR135 pKa = 11.84TMDD138 pKa = 2.98HH139 pKa = 6.79AGLMGALSNEE149 pKa = 3.24KK150 pKa = 10.07FMYY153 pKa = 9.62FKK155 pKa = 10.11PVWMKK160 pKa = 9.8TIFKK164 pKa = 10.53VRR166 pKa = 11.84VTGGADD172 pKa = 3.42PVWTCEE178 pKa = 3.83TMGRR182 pKa = 11.84KK183 pKa = 8.87CEE185 pKa = 4.0YY186 pKa = 10.06SSSGLPDD193 pKa = 4.97PYY195 pKa = 10.42TEE197 pKa = 4.83SKK199 pKa = 10.65IEE201 pKa = 3.97TDD203 pKa = 4.34FKK205 pKa = 11.1TKK207 pKa = 9.7MSFYY211 pKa = 10.77KK212 pKa = 10.34ILTAGKK218 pKa = 7.16TCKK221 pKa = 9.16YY222 pKa = 9.29TLFSRR227 pKa = 11.84LGEE230 pKa = 4.18NKK232 pKa = 10.38SIGPGSLQNPAATLISNYY250 pKa = 9.41IKK252 pKa = 10.44PSQWLLWNEE261 pKa = 4.85TIQNMGNWLNYY272 pKa = 8.97PVHH275 pKa = 6.25QLIVKK280 pKa = 9.83SGSFGIGAVDD290 pKa = 3.82AQRR293 pKa = 11.84MGLYY297 pKa = 9.87VEE299 pKa = 5.94AYY301 pKa = 7.75HH302 pKa = 6.58TVHH305 pKa = 7.02CLVAKK310 pKa = 10.68NNAVTQAYY318 pKa = 7.35NTAA321 pKa = 3.61
MM1 pKa = 7.51ARR3 pKa = 11.84RR4 pKa = 11.84GYY6 pKa = 10.1RR7 pKa = 11.84YY8 pKa = 9.73KK9 pKa = 10.53RR10 pKa = 11.84YY11 pKa = 9.38YY12 pKa = 9.85GKK14 pKa = 9.86NRR16 pKa = 11.84KK17 pKa = 9.07RR18 pKa = 11.84FIGPLLPGGYY28 pKa = 10.32AKK30 pKa = 10.93GNAKK34 pKa = 10.07SLARR38 pKa = 11.84KK39 pKa = 6.58WRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84YY45 pKa = 9.89RR46 pKa = 11.84YY47 pKa = 9.48GKK49 pKa = 10.01KK50 pKa = 9.92SKK52 pKa = 10.37QSLRR56 pKa = 11.84SGWGLGMTRR65 pKa = 11.84AKK67 pKa = 10.75SFVKK71 pKa = 10.32RR72 pKa = 11.84VDD74 pKa = 3.86LNEE77 pKa = 3.93FTPKK81 pKa = 9.98GSCRR85 pKa = 11.84FRR87 pKa = 11.84ISYY90 pKa = 10.1RR91 pKa = 11.84GRR93 pKa = 11.84HH94 pKa = 6.51LITAANVYY102 pKa = 9.68CAANFHH108 pKa = 6.22PHH110 pKa = 5.4PHH112 pKa = 5.94YY113 pKa = 8.05PTTGFADD120 pKa = 3.86PVLSATSDD128 pKa = 3.58DD129 pKa = 3.98FTSDD133 pKa = 3.35PRR135 pKa = 11.84TMDD138 pKa = 2.98HH139 pKa = 6.79AGLMGALSNEE149 pKa = 3.24KK150 pKa = 10.07FMYY153 pKa = 9.62FKK155 pKa = 10.11PVWMKK160 pKa = 9.8TIFKK164 pKa = 10.53VRR166 pKa = 11.84VTGGADD172 pKa = 3.42PVWTCEE178 pKa = 3.83TMGRR182 pKa = 11.84KK183 pKa = 8.87CEE185 pKa = 4.0YY186 pKa = 10.06SSSGLPDD193 pKa = 4.97PYY195 pKa = 10.42TEE197 pKa = 4.83SKK199 pKa = 10.65IEE201 pKa = 3.97TDD203 pKa = 4.34FKK205 pKa = 11.1TKK207 pKa = 9.7MSFYY211 pKa = 10.77KK212 pKa = 10.34ILTAGKK218 pKa = 7.16TCKK221 pKa = 9.16YY222 pKa = 9.29TLFSRR227 pKa = 11.84LGEE230 pKa = 4.18NKK232 pKa = 10.38SIGPGSLQNPAATLISNYY250 pKa = 9.41IKK252 pKa = 10.44PSQWLLWNEE261 pKa = 4.85TIQNMGNWLNYY272 pKa = 8.97PVHH275 pKa = 6.25QLIVKK280 pKa = 9.83SGSFGIGAVDD290 pKa = 3.82AQRR293 pKa = 11.84MGLYY297 pKa = 9.87VEE299 pKa = 5.94AYY301 pKa = 7.75HH302 pKa = 6.58TVHH305 pKa = 7.02CLVAKK310 pKa = 10.68NNAVTQAYY318 pKa = 7.35NTAA321 pKa = 3.61
Molecular weight: 36.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
632 |
311 |
321 |
316.0 |
36.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.171 ± 0.923 | 2.215 ± 0.245 |
4.272 ± 0.818 | 5.38 ± 1.821 |
3.956 ± 0.287 | 7.911 ± 0.574 |
2.848 ± 0.252 | 5.063 ± 0.937 |
6.487 ± 1.36 | 6.962 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.848 ± 0.189 | 4.114 ± 0.615 |
4.905 ± 0.056 | 3.639 ± 1.031 |
7.911 ± 0.307 | 6.013 ± 0.815 |
6.804 ± 0.255 | 4.589 ± 0.06 |
2.373 ± 0.136 | 5.538 ± 0.49 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
