
Clostridium sp. CAG:510
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
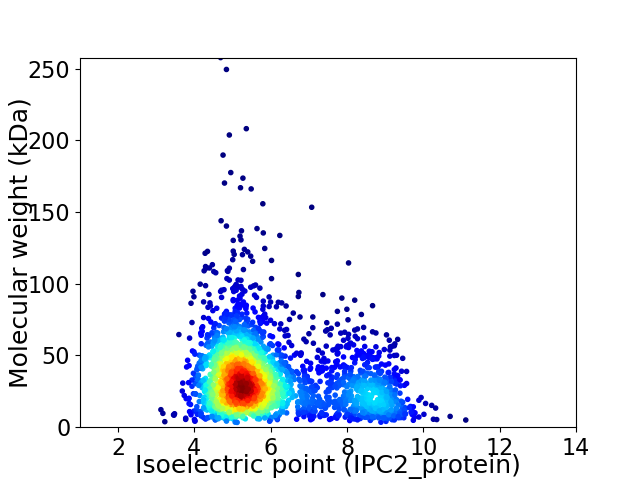
Virtual 2D-PAGE plot for 2541 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6C5D9|R6C5D9_9CLOT MATE efflux family protein OS=Clostridium sp. CAG:510 OX=1262816 GN=BN687_00327 PE=4 SV=1
MM1 pKa = 7.25KK2 pKa = 10.51KK3 pKa = 10.1KK4 pKa = 9.48MKK6 pKa = 10.43RR7 pKa = 11.84FFASVLAFAMMFSTLTFTSAAAEE30 pKa = 4.32DD31 pKa = 4.67EE32 pKa = 4.46YY33 pKa = 11.77QMFLAIGADD42 pKa = 3.59AEE44 pKa = 4.55ADD46 pKa = 3.82GDD48 pKa = 3.49WGYY51 pKa = 11.37QYY53 pKa = 10.87YY54 pKa = 11.27GNGASSNAGDD64 pKa = 3.84VTATDD69 pKa = 3.56ATAKK73 pKa = 10.79VGDD76 pKa = 4.01TVTIGVSFPTEE87 pKa = 3.87VVYY90 pKa = 9.19TWFMAPVLVAEE101 pKa = 4.39NAGNVDD107 pKa = 3.58YY108 pKa = 10.45TIDD111 pKa = 3.47SVKK114 pKa = 10.12IDD116 pKa = 4.02GEE118 pKa = 4.36DD119 pKa = 3.38VTDD122 pKa = 6.07KK123 pKa = 11.0IDD125 pKa = 3.7LSAGKK130 pKa = 10.11AFWYY134 pKa = 9.87EE135 pKa = 3.73GTGDD139 pKa = 3.73YY140 pKa = 11.23SEE142 pKa = 4.41TQAVRR147 pKa = 11.84LAGGYY152 pKa = 8.77NEE154 pKa = 4.8WGDD157 pKa = 3.56KK158 pKa = 10.64YY159 pKa = 10.45MAEE162 pKa = 4.1SPKK165 pKa = 10.29GFKK168 pKa = 10.36EE169 pKa = 3.42ITYY172 pKa = 8.89TITLNAVEE180 pKa = 4.58EE181 pKa = 4.5GAAAGEE187 pKa = 4.24ATLSEE192 pKa = 4.21EE193 pKa = 4.79SYY195 pKa = 11.32DD196 pKa = 3.58AFIAIGADD204 pKa = 3.36KK205 pKa = 10.46EE206 pKa = 4.42AEE208 pKa = 4.04NDD210 pKa = 2.77WGYY213 pKa = 11.81NYY215 pKa = 10.58AGEE218 pKa = 4.24DD219 pKa = 3.68AEE221 pKa = 5.81GITATTGTLKK231 pKa = 10.78SGEE234 pKa = 4.34TTTLSLEE241 pKa = 3.88FDD243 pKa = 3.68SPVFYY248 pKa = 9.47TWYY251 pKa = 9.88VAPCMVVDD259 pKa = 4.41DD260 pKa = 5.43ASAISDD266 pKa = 3.22QSTFDD271 pKa = 3.37VKK273 pKa = 11.2VYY275 pKa = 10.95LDD277 pKa = 4.22DD278 pKa = 5.12EE279 pKa = 4.65EE280 pKa = 4.71VTTDD284 pKa = 3.45LSAGKK289 pKa = 9.9VCWAEE294 pKa = 3.67GTGAYY299 pKa = 10.31DD300 pKa = 3.15EE301 pKa = 4.8TKK303 pKa = 10.32CIRR306 pKa = 11.84IGGGYY311 pKa = 9.08NEE313 pKa = 4.87WGDD316 pKa = 3.59KK317 pKa = 10.45YY318 pKa = 10.95LAEE321 pKa = 4.34SPKK324 pKa = 10.27GYY326 pKa = 10.6KK327 pKa = 10.13KK328 pKa = 9.16ITFEE332 pKa = 4.16ITPEE336 pKa = 3.87IYY338 pKa = 10.04VAAAAEE344 pKa = 4.32EE345 pKa = 4.61TVTVAPVDD353 pKa = 4.29LNGTYY358 pKa = 10.28NAYY361 pKa = 9.76IGIQTPNWSFRR372 pKa = 11.84NAYY375 pKa = 10.33DD376 pKa = 3.59DD377 pKa = 3.48ATYY380 pKa = 11.39GLGTDD385 pKa = 4.99FFNQITGWDD394 pKa = 3.79ADD396 pKa = 3.94NNAITIPGNIHH407 pKa = 6.38DD408 pKa = 4.62TVISGNGTYY417 pKa = 9.24TVSIDD422 pKa = 3.45GLSFPDD428 pKa = 4.74GEE430 pKa = 4.81FSTQEE435 pKa = 3.79YY436 pKa = 10.37LNLIFISTDD445 pKa = 2.73IPNSGEE451 pKa = 3.76ITFSDD456 pKa = 3.5VKK458 pKa = 10.54LTIDD462 pKa = 4.4GKK464 pKa = 10.63NVDD467 pKa = 4.59LPNGPVLDD475 pKa = 4.71PDD477 pKa = 3.91SKK479 pKa = 11.31EE480 pKa = 3.97VTKK483 pKa = 11.09LLLQNIWNNDD493 pKa = 3.34CKK495 pKa = 10.94EE496 pKa = 3.72IGYY499 pKa = 9.35YY500 pKa = 9.52AVPMTDD506 pKa = 2.47ISITFTVSGFAYY518 pKa = 10.37DD519 pKa = 4.01NEE521 pKa = 4.36ASAEE525 pKa = 4.21TPEE528 pKa = 4.48TVAEE532 pKa = 4.14SAPAEE537 pKa = 4.16TTPAAEE543 pKa = 4.28EE544 pKa = 4.24TSSNTGLIVGIVVAAVVVIGGVVAGVVVSKK574 pKa = 10.69KK575 pKa = 10.44KK576 pKa = 10.13KK577 pKa = 10.13ANN579 pKa = 3.4
MM1 pKa = 7.25KK2 pKa = 10.51KK3 pKa = 10.1KK4 pKa = 9.48MKK6 pKa = 10.43RR7 pKa = 11.84FFASVLAFAMMFSTLTFTSAAAEE30 pKa = 4.32DD31 pKa = 4.67EE32 pKa = 4.46YY33 pKa = 11.77QMFLAIGADD42 pKa = 3.59AEE44 pKa = 4.55ADD46 pKa = 3.82GDD48 pKa = 3.49WGYY51 pKa = 11.37QYY53 pKa = 10.87YY54 pKa = 11.27GNGASSNAGDD64 pKa = 3.84VTATDD69 pKa = 3.56ATAKK73 pKa = 10.79VGDD76 pKa = 4.01TVTIGVSFPTEE87 pKa = 3.87VVYY90 pKa = 9.19TWFMAPVLVAEE101 pKa = 4.39NAGNVDD107 pKa = 3.58YY108 pKa = 10.45TIDD111 pKa = 3.47SVKK114 pKa = 10.12IDD116 pKa = 4.02GEE118 pKa = 4.36DD119 pKa = 3.38VTDD122 pKa = 6.07KK123 pKa = 11.0IDD125 pKa = 3.7LSAGKK130 pKa = 10.11AFWYY134 pKa = 9.87EE135 pKa = 3.73GTGDD139 pKa = 3.73YY140 pKa = 11.23SEE142 pKa = 4.41TQAVRR147 pKa = 11.84LAGGYY152 pKa = 8.77NEE154 pKa = 4.8WGDD157 pKa = 3.56KK158 pKa = 10.64YY159 pKa = 10.45MAEE162 pKa = 4.1SPKK165 pKa = 10.29GFKK168 pKa = 10.36EE169 pKa = 3.42ITYY172 pKa = 8.89TITLNAVEE180 pKa = 4.58EE181 pKa = 4.5GAAAGEE187 pKa = 4.24ATLSEE192 pKa = 4.21EE193 pKa = 4.79SYY195 pKa = 11.32DD196 pKa = 3.58AFIAIGADD204 pKa = 3.36KK205 pKa = 10.46EE206 pKa = 4.42AEE208 pKa = 4.04NDD210 pKa = 2.77WGYY213 pKa = 11.81NYY215 pKa = 10.58AGEE218 pKa = 4.24DD219 pKa = 3.68AEE221 pKa = 5.81GITATTGTLKK231 pKa = 10.78SGEE234 pKa = 4.34TTTLSLEE241 pKa = 3.88FDD243 pKa = 3.68SPVFYY248 pKa = 9.47TWYY251 pKa = 9.88VAPCMVVDD259 pKa = 4.41DD260 pKa = 5.43ASAISDD266 pKa = 3.22QSTFDD271 pKa = 3.37VKK273 pKa = 11.2VYY275 pKa = 10.95LDD277 pKa = 4.22DD278 pKa = 5.12EE279 pKa = 4.65EE280 pKa = 4.71VTTDD284 pKa = 3.45LSAGKK289 pKa = 9.9VCWAEE294 pKa = 3.67GTGAYY299 pKa = 10.31DD300 pKa = 3.15EE301 pKa = 4.8TKK303 pKa = 10.32CIRR306 pKa = 11.84IGGGYY311 pKa = 9.08NEE313 pKa = 4.87WGDD316 pKa = 3.59KK317 pKa = 10.45YY318 pKa = 10.95LAEE321 pKa = 4.34SPKK324 pKa = 10.27GYY326 pKa = 10.6KK327 pKa = 10.13KK328 pKa = 9.16ITFEE332 pKa = 4.16ITPEE336 pKa = 3.87IYY338 pKa = 10.04VAAAAEE344 pKa = 4.32EE345 pKa = 4.61TVTVAPVDD353 pKa = 4.29LNGTYY358 pKa = 10.28NAYY361 pKa = 9.76IGIQTPNWSFRR372 pKa = 11.84NAYY375 pKa = 10.33DD376 pKa = 3.59DD377 pKa = 3.48ATYY380 pKa = 11.39GLGTDD385 pKa = 4.99FFNQITGWDD394 pKa = 3.79ADD396 pKa = 3.94NNAITIPGNIHH407 pKa = 6.38DD408 pKa = 4.62TVISGNGTYY417 pKa = 9.24TVSIDD422 pKa = 3.45GLSFPDD428 pKa = 4.74GEE430 pKa = 4.81FSTQEE435 pKa = 3.79YY436 pKa = 10.37LNLIFISTDD445 pKa = 2.73IPNSGEE451 pKa = 3.76ITFSDD456 pKa = 3.5VKK458 pKa = 10.54LTIDD462 pKa = 4.4GKK464 pKa = 10.63NVDD467 pKa = 4.59LPNGPVLDD475 pKa = 4.71PDD477 pKa = 3.91SKK479 pKa = 11.31EE480 pKa = 3.97VTKK483 pKa = 11.09LLLQNIWNNDD493 pKa = 3.34CKK495 pKa = 10.94EE496 pKa = 3.72IGYY499 pKa = 9.35YY500 pKa = 9.52AVPMTDD506 pKa = 2.47ISITFTVSGFAYY518 pKa = 10.37DD519 pKa = 4.01NEE521 pKa = 4.36ASAEE525 pKa = 4.21TPEE528 pKa = 4.48TVAEE532 pKa = 4.14SAPAEE537 pKa = 4.16TTPAAEE543 pKa = 4.28EE544 pKa = 4.24TSSNTGLIVGIVVAAVVVIGGVVAGVVVSKK574 pKa = 10.69KK575 pKa = 10.44KK576 pKa = 10.13KK577 pKa = 10.13ANN579 pKa = 3.4
Molecular weight: 62.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6C6X3|R6C6X3_9CLOT Phosphate acetyltransferase OS=Clostridium sp. CAG:510 OX=1262816 GN=BN687_00830 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.21GRR39 pKa = 11.84HH40 pKa = 5.51KK41 pKa = 10.9LSAA44 pKa = 3.8
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.21GRR39 pKa = 11.84HH40 pKa = 5.51KK41 pKa = 10.9LSAA44 pKa = 3.8
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
814344 |
29 |
2345 |
320.5 |
35.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.722 ± 0.057 | 1.473 ± 0.018 |
5.666 ± 0.037 | 7.917 ± 0.054 |
4.132 ± 0.031 | 7.174 ± 0.047 |
1.628 ± 0.021 | 6.998 ± 0.04 |
7.045 ± 0.038 | 8.937 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.117 ± 0.028 | 4.285 ± 0.033 |
3.237 ± 0.026 | 2.972 ± 0.026 |
4.246 ± 0.038 | 5.549 ± 0.034 |
5.645 ± 0.039 | 7.004 ± 0.041 |
0.871 ± 0.015 | 4.38 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
