
Bacteroides sp. CAG:633
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
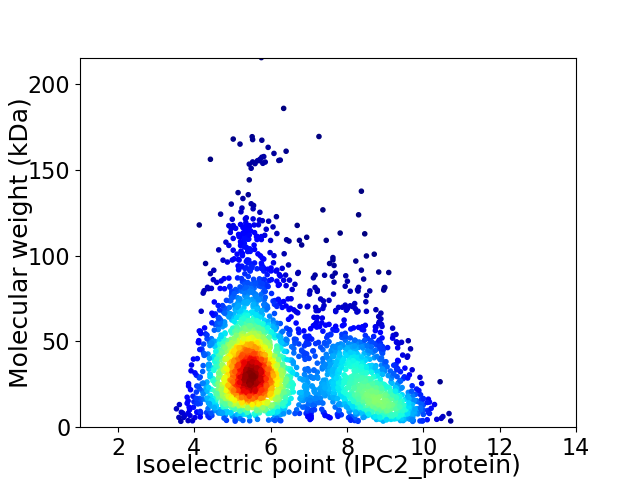
Virtual 2D-PAGE plot for 3004 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6FPN9|R6FPN9_9BACE Rhamnulokinase OS=Bacteroides sp. CAG:633 OX=1262744 GN=BN744_02636 PE=4 SV=1
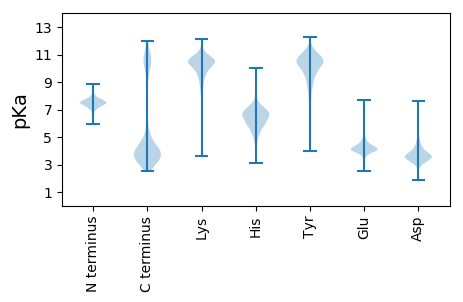
MM1 pKa = 6.94NTRR4 pKa = 11.84NVNIPDD10 pKa = 3.85IDD12 pKa = 4.8DD13 pKa = 4.79LDD15 pKa = 4.22ALCRR19 pKa = 11.84AIDD22 pKa = 3.41WTQYY26 pKa = 11.24QSMLEE31 pKa = 3.75QSLANEE37 pKa = 4.26KK38 pKa = 8.45TWEE41 pKa = 4.29FGCMDD46 pKa = 4.92EE47 pKa = 5.12YY48 pKa = 11.62NPHH51 pKa = 6.56TDD53 pKa = 3.15NIAQIEE59 pKa = 4.36EE60 pKa = 4.19EE61 pKa = 4.78LNFLSAGEE69 pKa = 4.12YY70 pKa = 8.51EE71 pKa = 5.1AIVRR75 pKa = 11.84MHH77 pKa = 6.86DD78 pKa = 2.96AEE80 pKa = 4.3YY81 pKa = 10.45FQDD84 pKa = 4.73FVV86 pKa = 3.82
MM1 pKa = 6.94NTRR4 pKa = 11.84NVNIPDD10 pKa = 3.85IDD12 pKa = 4.8DD13 pKa = 4.79LDD15 pKa = 4.22ALCRR19 pKa = 11.84AIDD22 pKa = 3.41WTQYY26 pKa = 11.24QSMLEE31 pKa = 3.75QSLANEE37 pKa = 4.26KK38 pKa = 8.45TWEE41 pKa = 4.29FGCMDD46 pKa = 4.92EE47 pKa = 5.12YY48 pKa = 11.62NPHH51 pKa = 6.56TDD53 pKa = 3.15NIAQIEE59 pKa = 4.36EE60 pKa = 4.19EE61 pKa = 4.78LNFLSAGEE69 pKa = 4.12YY70 pKa = 8.51EE71 pKa = 5.1AIVRR75 pKa = 11.84MHH77 pKa = 6.86DD78 pKa = 2.96AEE80 pKa = 4.3YY81 pKa = 10.45FQDD84 pKa = 4.73FVV86 pKa = 3.82
Molecular weight: 10.13 kDa
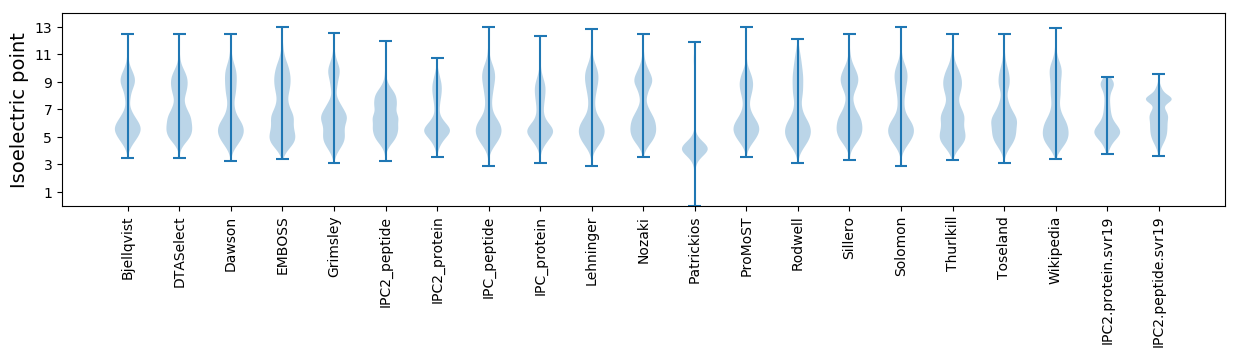
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6FBJ2|R6FBJ2_9BACE Transporter gate domain protein OS=Bacteroides sp. CAG:633 OX=1262744 GN=BN744_00813 PE=4 SV=1
MM1 pKa = 7.37EE2 pKa = 6.44PIRR5 pKa = 11.84NFDD8 pKa = 3.46QLTAHH13 pKa = 7.46LKK15 pKa = 8.81TLNKK19 pKa = 9.33RR20 pKa = 11.84KK21 pKa = 10.05RR22 pKa = 11.84IAVVCANDD30 pKa = 3.83PNTEE34 pKa = 4.08YY35 pKa = 10.81AISRR39 pKa = 11.84ALEE42 pKa = 3.65EE43 pKa = 5.14GIAEE47 pKa = 4.11FLMIGDD53 pKa = 3.91SAILEE58 pKa = 4.56KK59 pKa = 11.12YY60 pKa = 9.74PSLKK64 pKa = 10.53NYY66 pKa = 9.05PDD68 pKa = 3.84YY69 pKa = 11.69VHH71 pKa = 6.7TLHH74 pKa = 7.86VEE76 pKa = 4.41DD77 pKa = 5.01PDD79 pKa = 3.57EE80 pKa = 4.43AARR83 pKa = 11.84EE84 pKa = 3.88AVRR87 pKa = 11.84IVRR90 pKa = 11.84EE91 pKa = 4.18GGADD95 pKa = 2.97ILMKK99 pKa = 10.91GIINTDD105 pKa = 3.45NLLHH109 pKa = 7.4AILDD113 pKa = 4.01KK114 pKa = 11.34EE115 pKa = 4.46KK116 pKa = 11.14GLLPKK121 pKa = 10.77GKK123 pKa = 9.52ILTHH127 pKa = 6.22LAVMQIPTYY136 pKa = 10.94DD137 pKa = 3.29KK138 pKa = 11.43LLFFSDD144 pKa = 3.25AAVIPRR150 pKa = 11.84PTLQQRR156 pKa = 11.84IEE158 pKa = 4.28MIWYY162 pKa = 8.73AIRR165 pKa = 11.84ACRR168 pKa = 11.84HH169 pKa = 5.61FGIEE173 pKa = 3.72QPRR176 pKa = 11.84XLRR179 pKa = 11.84HH180 pKa = 5.6RR181 pKa = 11.84AAAHH185 pKa = 6.65RR186 pKa = 11.84PDD188 pKa = 3.37TLYY191 pKa = 11.36GEE193 pKa = 5.32GEE195 pKa = 4.08RR196 pKa = 11.84QVPPFAGLCEE206 pKa = 3.93HH207 pKa = 6.88RR208 pKa = 11.84RR209 pKa = 11.84TGRR212 pKa = 11.84SRR214 pKa = 11.84RR215 pKa = 11.84IRR217 pKa = 11.84QRR219 pKa = 11.84HH220 pKa = 4.44HH221 pKa = 6.34RR222 pKa = 11.84RR223 pKa = 11.84SAGRR227 pKa = 11.84EE228 pKa = 3.62NLVRR232 pKa = 11.84EE233 pKa = 4.41DD234 pKa = 3.31QRR236 pKa = 11.84RR237 pKa = 11.84HH238 pKa = 3.75QRR240 pKa = 11.84HH241 pKa = 5.48RR242 pKa = 11.84FAHH245 pKa = 4.46QRR247 pKa = 11.84RR248 pKa = 11.84GRR250 pKa = 11.84RR251 pKa = 11.84THH253 pKa = 7.03LPQHH257 pKa = 6.17RR258 pKa = 11.84VGQRR262 pKa = 11.84FLQSRR267 pKa = 11.84LPVLARR273 pKa = 11.84RR274 pKa = 11.84HH275 pKa = 4.76GRR277 pKa = 11.84IAARR281 pKa = 11.84PHH283 pKa = 6.09LSGGAALAQRR293 pKa = 11.84QRR295 pKa = 11.84TVEE298 pKa = 4.2VLQHH302 pKa = 5.59RR303 pKa = 11.84HH304 pKa = 5.51GLPDD308 pKa = 3.43EE309 pKa = 4.03QRR311 pKa = 11.84TGHH314 pKa = 5.76EE315 pKa = 4.33RR316 pKa = 11.84IKK318 pKa = 10.86QEE320 pKa = 3.63ASRR323 pKa = 11.84LTIHH327 pKa = 6.52HH328 pKa = 7.16LSFIIHH334 pKa = 6.76HH335 pKa = 5.44FHH337 pKa = 7.29EE338 pKa = 6.1DD339 pKa = 2.89ISHH342 pKa = 5.46QPRR345 pKa = 11.84LYY347 pKa = 10.54FDD349 pKa = 3.49EE350 pKa = 4.66DD351 pKa = 3.34CRR353 pKa = 11.84LRR355 pKa = 11.84RR356 pKa = 3.75
MM1 pKa = 7.37EE2 pKa = 6.44PIRR5 pKa = 11.84NFDD8 pKa = 3.46QLTAHH13 pKa = 7.46LKK15 pKa = 8.81TLNKK19 pKa = 9.33RR20 pKa = 11.84KK21 pKa = 10.05RR22 pKa = 11.84IAVVCANDD30 pKa = 3.83PNTEE34 pKa = 4.08YY35 pKa = 10.81AISRR39 pKa = 11.84ALEE42 pKa = 3.65EE43 pKa = 5.14GIAEE47 pKa = 4.11FLMIGDD53 pKa = 3.91SAILEE58 pKa = 4.56KK59 pKa = 11.12YY60 pKa = 9.74PSLKK64 pKa = 10.53NYY66 pKa = 9.05PDD68 pKa = 3.84YY69 pKa = 11.69VHH71 pKa = 6.7TLHH74 pKa = 7.86VEE76 pKa = 4.41DD77 pKa = 5.01PDD79 pKa = 3.57EE80 pKa = 4.43AARR83 pKa = 11.84EE84 pKa = 3.88AVRR87 pKa = 11.84IVRR90 pKa = 11.84EE91 pKa = 4.18GGADD95 pKa = 2.97ILMKK99 pKa = 10.91GIINTDD105 pKa = 3.45NLLHH109 pKa = 7.4AILDD113 pKa = 4.01KK114 pKa = 11.34EE115 pKa = 4.46KK116 pKa = 11.14GLLPKK121 pKa = 10.77GKK123 pKa = 9.52ILTHH127 pKa = 6.22LAVMQIPTYY136 pKa = 10.94DD137 pKa = 3.29KK138 pKa = 11.43LLFFSDD144 pKa = 3.25AAVIPRR150 pKa = 11.84PTLQQRR156 pKa = 11.84IEE158 pKa = 4.28MIWYY162 pKa = 8.73AIRR165 pKa = 11.84ACRR168 pKa = 11.84HH169 pKa = 5.61FGIEE173 pKa = 3.72QPRR176 pKa = 11.84XLRR179 pKa = 11.84HH180 pKa = 5.6RR181 pKa = 11.84AAAHH185 pKa = 6.65RR186 pKa = 11.84PDD188 pKa = 3.37TLYY191 pKa = 11.36GEE193 pKa = 5.32GEE195 pKa = 4.08RR196 pKa = 11.84QVPPFAGLCEE206 pKa = 3.93HH207 pKa = 6.88RR208 pKa = 11.84RR209 pKa = 11.84TGRR212 pKa = 11.84SRR214 pKa = 11.84RR215 pKa = 11.84IRR217 pKa = 11.84QRR219 pKa = 11.84HH220 pKa = 4.44HH221 pKa = 6.34RR222 pKa = 11.84RR223 pKa = 11.84SAGRR227 pKa = 11.84EE228 pKa = 3.62NLVRR232 pKa = 11.84EE233 pKa = 4.41DD234 pKa = 3.31QRR236 pKa = 11.84RR237 pKa = 11.84HH238 pKa = 3.75QRR240 pKa = 11.84HH241 pKa = 5.48RR242 pKa = 11.84FAHH245 pKa = 4.46QRR247 pKa = 11.84RR248 pKa = 11.84GRR250 pKa = 11.84RR251 pKa = 11.84THH253 pKa = 7.03LPQHH257 pKa = 6.17RR258 pKa = 11.84VGQRR262 pKa = 11.84FLQSRR267 pKa = 11.84LPVLARR273 pKa = 11.84RR274 pKa = 11.84HH275 pKa = 4.76GRR277 pKa = 11.84IAARR281 pKa = 11.84PHH283 pKa = 6.09LSGGAALAQRR293 pKa = 11.84QRR295 pKa = 11.84TVEE298 pKa = 4.2VLQHH302 pKa = 5.59RR303 pKa = 11.84HH304 pKa = 5.51GLPDD308 pKa = 3.43EE309 pKa = 4.03QRR311 pKa = 11.84TGHH314 pKa = 5.76EE315 pKa = 4.33RR316 pKa = 11.84IKK318 pKa = 10.86QEE320 pKa = 3.63ASRR323 pKa = 11.84LTIHH327 pKa = 6.52HH328 pKa = 7.16LSFIIHH334 pKa = 6.76HH335 pKa = 5.44FHH337 pKa = 7.29EE338 pKa = 6.1DD339 pKa = 2.89ISHH342 pKa = 5.46QPRR345 pKa = 11.84LYY347 pKa = 10.54FDD349 pKa = 3.49EE350 pKa = 4.66DD351 pKa = 3.34CRR353 pKa = 11.84LRR355 pKa = 11.84RR356 pKa = 3.75
Molecular weight: 41.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1032115 |
29 |
1910 |
343.6 |
38.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.514 ± 0.047 | 1.342 ± 0.017 |
5.622 ± 0.03 | 6.516 ± 0.043 |
4.447 ± 0.027 | 6.898 ± 0.04 |
1.989 ± 0.022 | 6.347 ± 0.053 |
6.021 ± 0.042 | 9.377 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.76 ± 0.02 | 4.771 ± 0.042 |
3.811 ± 0.027 | 3.718 ± 0.029 |
5.008 ± 0.038 | 5.875 ± 0.037 |
5.597 ± 0.033 | 6.674 ± 0.044 |
1.294 ± 0.019 | 4.415 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
