
Alpaca respiratory coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Duvinacovirus; Human coronavirus 229E
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
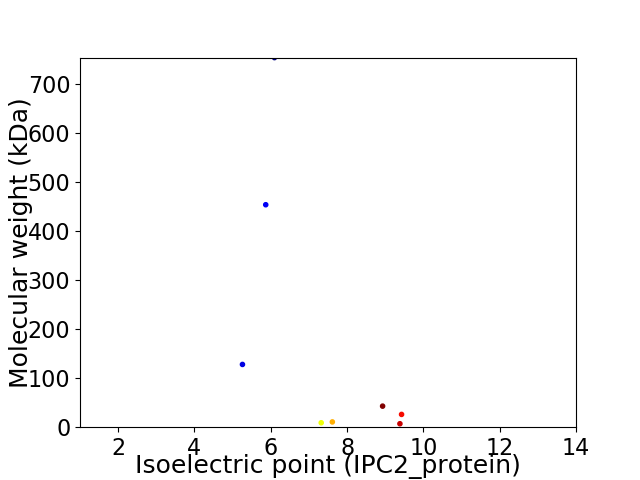
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1VWF6|I1VWF6_CVH22 Truncated ORF4 OS=Alpaca respiratory coronavirus OX=1176035 GN=ORF4 PE=4 SV=1
MM1 pKa = 7.09FVLVAYY7 pKa = 10.03ALLHH11 pKa = 6.13IAGCQTINVLNTSHH25 pKa = 6.59SVCNGCVGYY34 pKa = 10.09SEE36 pKa = 5.49NVFAVEE42 pKa = 4.01SGGYY46 pKa = 9.24IPSDD50 pKa = 3.14FAFNNWFLLTNTSSLVDD67 pKa = 3.51GVVRR71 pKa = 11.84SFQPLLLNCLWSVSGSRR88 pKa = 11.84FTTGFVYY95 pKa = 10.41FNGTGRR101 pKa = 11.84GDD103 pKa = 3.63CKK105 pKa = 10.87GFSSNVLSDD114 pKa = 3.61VIRR117 pKa = 11.84YY118 pKa = 8.25NINFEE123 pKa = 4.16EE124 pKa = 4.17NLRR127 pKa = 11.84RR128 pKa = 11.84GTILFKK134 pKa = 10.5TSFGVVVFYY143 pKa = 8.58CTNNTLVSGDD153 pKa = 3.46AYY155 pKa = 10.18IPFGTVLGNFYY166 pKa = 10.84CFVNTTIGNEE176 pKa = 4.22TTSSFVGALPKK187 pKa = 9.64TVRR190 pKa = 11.84EE191 pKa = 4.1FVISRR196 pKa = 11.84TGHH199 pKa = 6.13FYY201 pKa = 10.78INGYY205 pKa = 10.46RR206 pKa = 11.84YY207 pKa = 7.33FTLGNVEE214 pKa = 3.93AVNFNVTNAEE224 pKa = 4.21TTFCTVALASYY235 pKa = 11.29ADD237 pKa = 3.47VLVNVSQTAIANIIYY252 pKa = 9.57CNSVINRR259 pKa = 11.84LRR261 pKa = 11.84CDD263 pKa = 3.35QLSFDD268 pKa = 4.48VPDD271 pKa = 4.15GFYY274 pKa = 9.65STSPIQSVEE283 pKa = 4.08LPVSIVSLPVYY294 pKa = 9.8HH295 pKa = 6.85KK296 pKa = 9.52HH297 pKa = 4.94TFIVLYY303 pKa = 10.81VDD305 pKa = 5.71FKK307 pKa = 9.27PQSGAGTCYY316 pKa = 9.24NCRR319 pKa = 11.84PSVVNITLANFNEE332 pKa = 4.85TKK334 pKa = 10.76GPLCVEE340 pKa = 4.67TSHH343 pKa = 6.75FTTKK347 pKa = 10.41YY348 pKa = 7.97VASNVGRR355 pKa = 11.84WSASINTGNCPFSFGKK371 pKa = 10.56VNNFVKK377 pKa = 10.67FGSVCFSLKK386 pKa = 10.45DD387 pKa = 3.56IPGGCAMPIVANFAYY402 pKa = 9.7INSYY406 pKa = 9.54TIGSLYY412 pKa = 10.74VSWSDD417 pKa = 3.16GDD419 pKa = 4.55GITGVPKK426 pKa = 10.08PVEE429 pKa = 4.35GVSSFMNVTLNKK441 pKa = 7.67CTKK444 pKa = 9.28YY445 pKa = 10.78NIYY448 pKa = 10.19DD449 pKa = 3.42VSGVGVIRR457 pKa = 11.84ISNDD461 pKa = 2.59TFLNGITYY469 pKa = 9.16TSTSGNLLGFKK480 pKa = 10.35DD481 pKa = 3.72VTNGIIYY488 pKa = 10.18SITPCNPPDD497 pKa = 3.49QLVVYY502 pKa = 8.2QQAVVGAMLSEE513 pKa = 4.31NSSSYY518 pKa = 10.69GFSNVVEE525 pKa = 4.56LPNFFYY531 pKa = 10.92ASNGTYY537 pKa = 10.3NCTDD541 pKa = 2.86AVLTYY546 pKa = 11.05SSFGVCADD554 pKa = 3.43GSIIAVQPRR563 pKa = 11.84NVSYY567 pKa = 10.96DD568 pKa = 3.28GVSAIVTANLSIPSNWTTSVQVEE591 pKa = 4.12YY592 pKa = 10.83LQITSTPILVDD603 pKa = 3.61CSTYY607 pKa = 10.14VCNGNVRR614 pKa = 11.84CVEE617 pKa = 4.02LLKK620 pKa = 10.78QYY622 pKa = 10.53TSACKK627 pKa = 9.83TIEE630 pKa = 4.06DD631 pKa = 4.09ALRR634 pKa = 11.84ISAMLEE640 pKa = 3.94SADD643 pKa = 3.73VGEE646 pKa = 4.64MLTFDD651 pKa = 3.92EE652 pKa = 5.67KK653 pKa = 11.5AFTLANVSSFGDD665 pKa = 3.69YY666 pKa = 10.6NLSSVIPSLPTSGSRR681 pKa = 11.84VAGRR685 pKa = 11.84SAIEE689 pKa = 5.09DD690 pKa = 3.29ILFSKK695 pKa = 10.76VVTSGLGTVDD705 pKa = 4.5ADD707 pKa = 3.86YY708 pKa = 11.22KK709 pKa = 11.03KK710 pKa = 9.54CTKK713 pKa = 10.13GLSIADD719 pKa = 4.44LACAQYY725 pKa = 11.34YY726 pKa = 9.2NGIMVLPGVADD737 pKa = 4.01AEE739 pKa = 4.12RR740 pKa = 11.84MAMYY744 pKa = 9.17TGSLIGGIALGGLTSAAAIPFSLALQARR772 pKa = 11.84LNYY775 pKa = 9.74VALQTDD781 pKa = 4.22VLQEE785 pKa = 3.94NQKK788 pKa = 9.9ILAASFNKK796 pKa = 10.34AMTNIVDD803 pKa = 3.87AFTGVNDD810 pKa = 6.2AITQTSQAIQTVATALNKK828 pKa = 10.09IQDD831 pKa = 3.83VVNQQGNALNHH842 pKa = 5.93LTSQLRR848 pKa = 11.84QNFQAISSSIQAIYY862 pKa = 10.75DD863 pKa = 3.62RR864 pKa = 11.84LDD866 pKa = 3.71TIQADD871 pKa = 3.81QQVDD875 pKa = 3.53RR876 pKa = 11.84LITGRR881 pKa = 11.84LAALNAFVAQTLTKK895 pKa = 8.19YY896 pKa = 9.39TEE898 pKa = 4.13VRR900 pKa = 11.84ASRR903 pKa = 11.84QLAQQKK909 pKa = 10.1VNEE912 pKa = 4.46CVKK915 pKa = 10.52SQSNRR920 pKa = 11.84YY921 pKa = 7.84GFCGNGTHH929 pKa = 6.84IFSIVNSAPEE939 pKa = 4.07GLVFLHH945 pKa = 6.06TVLLPTQYY953 pKa = 11.19KK954 pKa = 10.38DD955 pKa = 3.27VEE957 pKa = 4.35AWSGLCVDD965 pKa = 3.87NTNGYY970 pKa = 7.62VLRR973 pKa = 11.84QPNLALYY980 pKa = 10.59KK981 pKa = 9.95EE982 pKa = 4.32GNYY985 pKa = 10.13YY986 pKa = 10.71RR987 pKa = 11.84ITSRR991 pKa = 11.84FMFEE995 pKa = 3.65PRR997 pKa = 11.84IPTMADD1003 pKa = 3.53FVQIEE1008 pKa = 4.38KK1009 pKa = 11.09CNVTFVNISRR1019 pKa = 11.84SEE1021 pKa = 3.96LQTIVPEE1028 pKa = 4.33YY1029 pKa = 10.5IDD1031 pKa = 3.57VNKK1034 pKa = 9.35TLQEE1038 pKa = 4.88LIDD1041 pKa = 4.09KK1042 pKa = 9.89LPNYY1046 pKa = 7.3TVPDD1050 pKa = 4.22LGIDD1054 pKa = 3.54QYY1056 pKa = 12.07NQTILNLTSEE1066 pKa = 4.16ISTLEE1071 pKa = 4.02NKK1073 pKa = 9.98SAEE1076 pKa = 4.14LNYY1079 pKa = 9.72TVQRR1083 pKa = 11.84LQTLIDD1089 pKa = 4.6NINSTLVDD1097 pKa = 4.5LKK1099 pKa = 10.17WLNRR1103 pKa = 11.84VEE1105 pKa = 5.47TYY1107 pKa = 10.68LKK1109 pKa = 9.22WPWWVWLCISVVLIFVVSILLLCCCSTGCCGFFSCLASSTRR1150 pKa = 11.84GCCEE1154 pKa = 3.91STKK1157 pKa = 10.6LPYY1160 pKa = 10.38YY1161 pKa = 10.29DD1162 pKa = 3.56VEE1164 pKa = 5.49KK1165 pKa = 10.86IHH1167 pKa = 6.47IQQ1169 pKa = 3.23
MM1 pKa = 7.09FVLVAYY7 pKa = 10.03ALLHH11 pKa = 6.13IAGCQTINVLNTSHH25 pKa = 6.59SVCNGCVGYY34 pKa = 10.09SEE36 pKa = 5.49NVFAVEE42 pKa = 4.01SGGYY46 pKa = 9.24IPSDD50 pKa = 3.14FAFNNWFLLTNTSSLVDD67 pKa = 3.51GVVRR71 pKa = 11.84SFQPLLLNCLWSVSGSRR88 pKa = 11.84FTTGFVYY95 pKa = 10.41FNGTGRR101 pKa = 11.84GDD103 pKa = 3.63CKK105 pKa = 10.87GFSSNVLSDD114 pKa = 3.61VIRR117 pKa = 11.84YY118 pKa = 8.25NINFEE123 pKa = 4.16EE124 pKa = 4.17NLRR127 pKa = 11.84RR128 pKa = 11.84GTILFKK134 pKa = 10.5TSFGVVVFYY143 pKa = 8.58CTNNTLVSGDD153 pKa = 3.46AYY155 pKa = 10.18IPFGTVLGNFYY166 pKa = 10.84CFVNTTIGNEE176 pKa = 4.22TTSSFVGALPKK187 pKa = 9.64TVRR190 pKa = 11.84EE191 pKa = 4.1FVISRR196 pKa = 11.84TGHH199 pKa = 6.13FYY201 pKa = 10.78INGYY205 pKa = 10.46RR206 pKa = 11.84YY207 pKa = 7.33FTLGNVEE214 pKa = 3.93AVNFNVTNAEE224 pKa = 4.21TTFCTVALASYY235 pKa = 11.29ADD237 pKa = 3.47VLVNVSQTAIANIIYY252 pKa = 9.57CNSVINRR259 pKa = 11.84LRR261 pKa = 11.84CDD263 pKa = 3.35QLSFDD268 pKa = 4.48VPDD271 pKa = 4.15GFYY274 pKa = 9.65STSPIQSVEE283 pKa = 4.08LPVSIVSLPVYY294 pKa = 9.8HH295 pKa = 6.85KK296 pKa = 9.52HH297 pKa = 4.94TFIVLYY303 pKa = 10.81VDD305 pKa = 5.71FKK307 pKa = 9.27PQSGAGTCYY316 pKa = 9.24NCRR319 pKa = 11.84PSVVNITLANFNEE332 pKa = 4.85TKK334 pKa = 10.76GPLCVEE340 pKa = 4.67TSHH343 pKa = 6.75FTTKK347 pKa = 10.41YY348 pKa = 7.97VASNVGRR355 pKa = 11.84WSASINTGNCPFSFGKK371 pKa = 10.56VNNFVKK377 pKa = 10.67FGSVCFSLKK386 pKa = 10.45DD387 pKa = 3.56IPGGCAMPIVANFAYY402 pKa = 9.7INSYY406 pKa = 9.54TIGSLYY412 pKa = 10.74VSWSDD417 pKa = 3.16GDD419 pKa = 4.55GITGVPKK426 pKa = 10.08PVEE429 pKa = 4.35GVSSFMNVTLNKK441 pKa = 7.67CTKK444 pKa = 9.28YY445 pKa = 10.78NIYY448 pKa = 10.19DD449 pKa = 3.42VSGVGVIRR457 pKa = 11.84ISNDD461 pKa = 2.59TFLNGITYY469 pKa = 9.16TSTSGNLLGFKK480 pKa = 10.35DD481 pKa = 3.72VTNGIIYY488 pKa = 10.18SITPCNPPDD497 pKa = 3.49QLVVYY502 pKa = 8.2QQAVVGAMLSEE513 pKa = 4.31NSSSYY518 pKa = 10.69GFSNVVEE525 pKa = 4.56LPNFFYY531 pKa = 10.92ASNGTYY537 pKa = 10.3NCTDD541 pKa = 2.86AVLTYY546 pKa = 11.05SSFGVCADD554 pKa = 3.43GSIIAVQPRR563 pKa = 11.84NVSYY567 pKa = 10.96DD568 pKa = 3.28GVSAIVTANLSIPSNWTTSVQVEE591 pKa = 4.12YY592 pKa = 10.83LQITSTPILVDD603 pKa = 3.61CSTYY607 pKa = 10.14VCNGNVRR614 pKa = 11.84CVEE617 pKa = 4.02LLKK620 pKa = 10.78QYY622 pKa = 10.53TSACKK627 pKa = 9.83TIEE630 pKa = 4.06DD631 pKa = 4.09ALRR634 pKa = 11.84ISAMLEE640 pKa = 3.94SADD643 pKa = 3.73VGEE646 pKa = 4.64MLTFDD651 pKa = 3.92EE652 pKa = 5.67KK653 pKa = 11.5AFTLANVSSFGDD665 pKa = 3.69YY666 pKa = 10.6NLSSVIPSLPTSGSRR681 pKa = 11.84VAGRR685 pKa = 11.84SAIEE689 pKa = 5.09DD690 pKa = 3.29ILFSKK695 pKa = 10.76VVTSGLGTVDD705 pKa = 4.5ADD707 pKa = 3.86YY708 pKa = 11.22KK709 pKa = 11.03KK710 pKa = 9.54CTKK713 pKa = 10.13GLSIADD719 pKa = 4.44LACAQYY725 pKa = 11.34YY726 pKa = 9.2NGIMVLPGVADD737 pKa = 4.01AEE739 pKa = 4.12RR740 pKa = 11.84MAMYY744 pKa = 9.17TGSLIGGIALGGLTSAAAIPFSLALQARR772 pKa = 11.84LNYY775 pKa = 9.74VALQTDD781 pKa = 4.22VLQEE785 pKa = 3.94NQKK788 pKa = 9.9ILAASFNKK796 pKa = 10.34AMTNIVDD803 pKa = 3.87AFTGVNDD810 pKa = 6.2AITQTSQAIQTVATALNKK828 pKa = 10.09IQDD831 pKa = 3.83VVNQQGNALNHH842 pKa = 5.93LTSQLRR848 pKa = 11.84QNFQAISSSIQAIYY862 pKa = 10.75DD863 pKa = 3.62RR864 pKa = 11.84LDD866 pKa = 3.71TIQADD871 pKa = 3.81QQVDD875 pKa = 3.53RR876 pKa = 11.84LITGRR881 pKa = 11.84LAALNAFVAQTLTKK895 pKa = 8.19YY896 pKa = 9.39TEE898 pKa = 4.13VRR900 pKa = 11.84ASRR903 pKa = 11.84QLAQQKK909 pKa = 10.1VNEE912 pKa = 4.46CVKK915 pKa = 10.52SQSNRR920 pKa = 11.84YY921 pKa = 7.84GFCGNGTHH929 pKa = 6.84IFSIVNSAPEE939 pKa = 4.07GLVFLHH945 pKa = 6.06TVLLPTQYY953 pKa = 11.19KK954 pKa = 10.38DD955 pKa = 3.27VEE957 pKa = 4.35AWSGLCVDD965 pKa = 3.87NTNGYY970 pKa = 7.62VLRR973 pKa = 11.84QPNLALYY980 pKa = 10.59KK981 pKa = 9.95EE982 pKa = 4.32GNYY985 pKa = 10.13YY986 pKa = 10.71RR987 pKa = 11.84ITSRR991 pKa = 11.84FMFEE995 pKa = 3.65PRR997 pKa = 11.84IPTMADD1003 pKa = 3.53FVQIEE1008 pKa = 4.38KK1009 pKa = 11.09CNVTFVNISRR1019 pKa = 11.84SEE1021 pKa = 3.96LQTIVPEE1028 pKa = 4.33YY1029 pKa = 10.5IDD1031 pKa = 3.57VNKK1034 pKa = 9.35TLQEE1038 pKa = 4.88LIDD1041 pKa = 4.09KK1042 pKa = 9.89LPNYY1046 pKa = 7.3TVPDD1050 pKa = 4.22LGIDD1054 pKa = 3.54QYY1056 pKa = 12.07NQTILNLTSEE1066 pKa = 4.16ISTLEE1071 pKa = 4.02NKK1073 pKa = 9.98SAEE1076 pKa = 4.14LNYY1079 pKa = 9.72TVQRR1083 pKa = 11.84LQTLIDD1089 pKa = 4.6NINSTLVDD1097 pKa = 4.5LKK1099 pKa = 10.17WLNRR1103 pKa = 11.84VEE1105 pKa = 5.47TYY1107 pKa = 10.68LKK1109 pKa = 9.22WPWWVWLCISVVLIFVVSILLLCCCSTGCCGFFSCLASSTRR1150 pKa = 11.84GCCEE1154 pKa = 3.91STKK1157 pKa = 10.6LPYY1160 pKa = 10.38YY1161 pKa = 10.29DD1162 pKa = 3.56VEE1164 pKa = 5.49KK1165 pKa = 10.86IHH1167 pKa = 6.47IQQ1169 pKa = 3.23
Molecular weight: 127.9 kDa
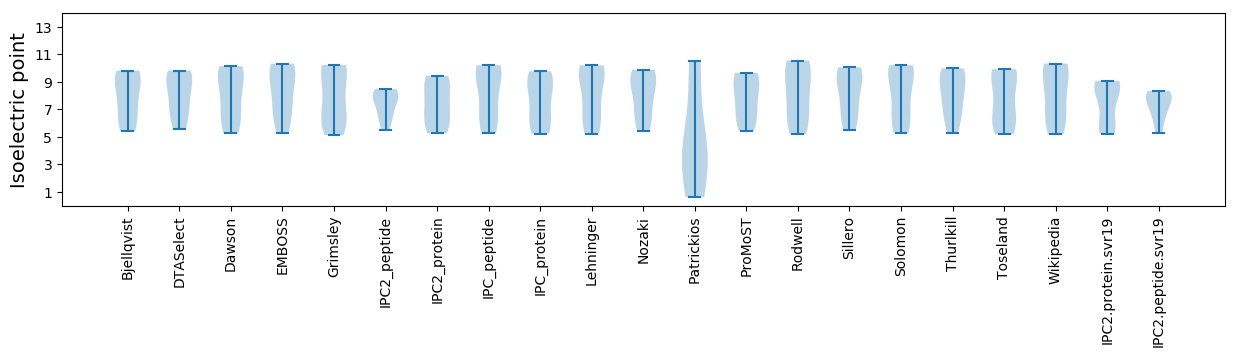
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1VWF7|I1VWF7_CVH22 Envelope small membrane protein OS=Alpaca respiratory coronavirus OX=1176035 GN=E PE=3 SV=1
MM1 pKa = 7.62ALGLFTLQIEE11 pKa = 4.69SAVNQSLSKK20 pKa = 10.41SKK22 pKa = 10.65VSAVVSRR29 pKa = 11.84QVIQDD34 pKa = 3.1VRR36 pKa = 11.84AAAVTFNLLAYY47 pKa = 8.89TLMSLFVVYY56 pKa = 9.16FAFFQSKK63 pKa = 8.24II64 pKa = 3.68
MM1 pKa = 7.62ALGLFTLQIEE11 pKa = 4.69SAVNQSLSKK20 pKa = 10.41SKK22 pKa = 10.65VSAVVSRR29 pKa = 11.84QVIQDD34 pKa = 3.1VRR36 pKa = 11.84AAAVTFNLLAYY47 pKa = 8.89TLMSLFVVYY56 pKa = 9.16FAFFQSKK63 pKa = 8.24II64 pKa = 3.68
Molecular weight: 7.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12866 |
64 |
6762 |
1608.3 |
178.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.917 ± 0.169 | 3.311 ± 0.396 |
5.41 ± 0.443 | 4.205 ± 0.155 |
5.689 ± 0.217 | 6.451 ± 0.135 |
1.687 ± 0.127 | 5.099 ± 0.254 |
6.008 ± 0.515 | 8.651 ± 0.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.153 ± 0.224 | 5.682 ± 0.283 |
3.397 ± 0.39 | 2.899 ± 0.413 |
3.257 ± 0.313 | 7.205 ± 0.559 |
5.938 ± 0.415 | 10.353 ± 0.323 |
1.36 ± 0.136 | 4.329 ± 0.207 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
