
Rhodococcus sp. OK302
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus; unclassified Rhodococcus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
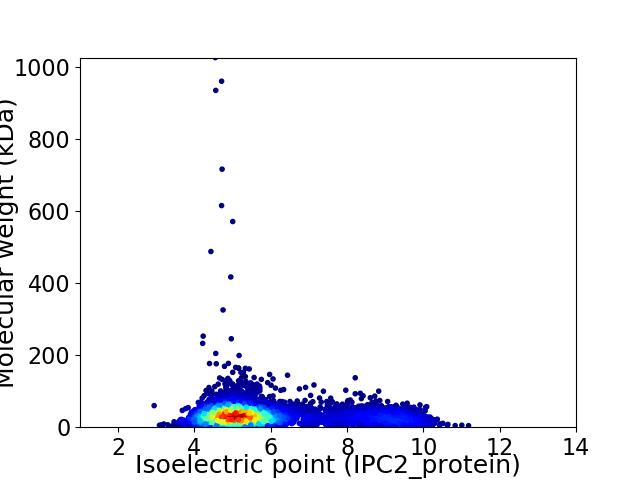
Virtual 2D-PAGE plot for 6148 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A235GA69|A0A235GA69_9NOCA Uncharacterized protein (TIGR02680 family) OS=Rhodococcus sp. OK302 OX=1882769 GN=BDB13_4089 PE=4 SV=1
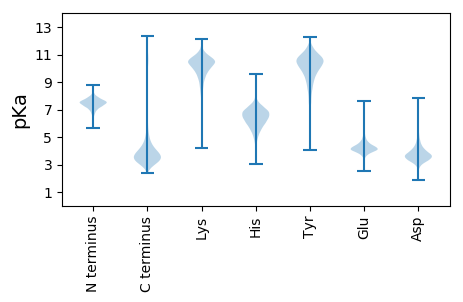
MM1 pKa = 7.18TNLYY5 pKa = 8.61RR6 pKa = 11.84TGRR9 pKa = 11.84GHH11 pKa = 6.06WRR13 pKa = 11.84SGLVAAILLVLAGCSASPSEE33 pKa = 4.59TEE35 pKa = 4.15SADD38 pKa = 3.44PAGPLPLGDD47 pKa = 3.55FTYY50 pKa = 10.88AANPCLDD57 pKa = 4.01PLIPGAPLPPFGDD70 pKa = 4.27DD71 pKa = 3.79FTCGTLIVPQNRR83 pKa = 11.84SQPGGATITIPVARR97 pKa = 11.84QRR99 pKa = 11.84AQNPQSEE106 pKa = 4.56MPPLLMLSGGPGGSGLLDD124 pKa = 3.43GLISYY129 pKa = 10.42SGLDD133 pKa = 3.54LNKK136 pKa = 10.45DD137 pKa = 3.0RR138 pKa = 11.84DD139 pKa = 4.0VIYY142 pKa = 10.16IDD144 pKa = 3.45QRR146 pKa = 11.84GTLRR150 pKa = 11.84ATPFVACPQVDD161 pKa = 3.25AVMQSLLDD169 pKa = 3.67KK170 pKa = 10.37PYY172 pKa = 10.37TDD174 pKa = 4.58PATEE178 pKa = 4.04AAGTAAVAACSEE190 pKa = 4.14QFRR193 pKa = 11.84AQGVDD198 pKa = 3.11LDD200 pKa = 4.26VFNSLEE206 pKa = 3.97NAADD210 pKa = 3.87LASLRR215 pKa = 11.84LALEE219 pKa = 3.9IPEE222 pKa = 3.91WDD224 pKa = 3.65VYY226 pKa = 10.2GVSYY230 pKa = 11.02GSDD233 pKa = 3.0LALQYY238 pKa = 11.19LRR240 pKa = 11.84DD241 pKa = 3.94FPDD244 pKa = 5.11GIRR247 pKa = 11.84AVVADD252 pKa = 4.17SVVPPQMNLADD263 pKa = 4.23TLWPNAARR271 pKa = 11.84GFDD274 pKa = 3.82ALEE277 pKa = 4.43AACDD281 pKa = 3.83AQPACQAMLPDD292 pKa = 4.02LTGTLATTVTDD303 pKa = 4.93LDD305 pKa = 3.47AHH307 pKa = 5.86PQVVSVTAGDD317 pKa = 4.21GSPVDD322 pKa = 3.94VMVDD326 pKa = 3.78GYY328 pKa = 11.72KK329 pKa = 10.37LASLVNSASLATNGLVDD346 pKa = 4.15MPAIIAAAGSGDD358 pKa = 3.82VTPAAQLLGSAQPSTVPLVGSGLTYY383 pKa = 10.84GVFCGEE389 pKa = 4.14AVAHH393 pKa = 6.23TSAEE397 pKa = 4.09AMFGAAKK404 pKa = 10.43NSLPQFPDD412 pKa = 2.97AVLSLTPQLPWLVGDD427 pKa = 4.31CAAWNVDD434 pKa = 3.53AVPDD438 pKa = 4.69LVVQPANSTVPVLLLSGGLDD458 pKa = 3.35GVTPPGNADD467 pKa = 3.01IAKK470 pKa = 8.2ATLPNSVALTFPEE483 pKa = 4.55SGHH486 pKa = 6.39GVLTQSDD493 pKa = 4.44CGPAAVTGFLDD504 pKa = 5.04DD505 pKa = 4.38PSASYY510 pKa = 10.63RR511 pKa = 11.84PQCLDD516 pKa = 3.35TVVIPPFSTSLVGSGG531 pKa = 3.6
MM1 pKa = 7.18TNLYY5 pKa = 8.61RR6 pKa = 11.84TGRR9 pKa = 11.84GHH11 pKa = 6.06WRR13 pKa = 11.84SGLVAAILLVLAGCSASPSEE33 pKa = 4.59TEE35 pKa = 4.15SADD38 pKa = 3.44PAGPLPLGDD47 pKa = 3.55FTYY50 pKa = 10.88AANPCLDD57 pKa = 4.01PLIPGAPLPPFGDD70 pKa = 4.27DD71 pKa = 3.79FTCGTLIVPQNRR83 pKa = 11.84SQPGGATITIPVARR97 pKa = 11.84QRR99 pKa = 11.84AQNPQSEE106 pKa = 4.56MPPLLMLSGGPGGSGLLDD124 pKa = 3.43GLISYY129 pKa = 10.42SGLDD133 pKa = 3.54LNKK136 pKa = 10.45DD137 pKa = 3.0RR138 pKa = 11.84DD139 pKa = 4.0VIYY142 pKa = 10.16IDD144 pKa = 3.45QRR146 pKa = 11.84GTLRR150 pKa = 11.84ATPFVACPQVDD161 pKa = 3.25AVMQSLLDD169 pKa = 3.67KK170 pKa = 10.37PYY172 pKa = 10.37TDD174 pKa = 4.58PATEE178 pKa = 4.04AAGTAAVAACSEE190 pKa = 4.14QFRR193 pKa = 11.84AQGVDD198 pKa = 3.11LDD200 pKa = 4.26VFNSLEE206 pKa = 3.97NAADD210 pKa = 3.87LASLRR215 pKa = 11.84LALEE219 pKa = 3.9IPEE222 pKa = 3.91WDD224 pKa = 3.65VYY226 pKa = 10.2GVSYY230 pKa = 11.02GSDD233 pKa = 3.0LALQYY238 pKa = 11.19LRR240 pKa = 11.84DD241 pKa = 3.94FPDD244 pKa = 5.11GIRR247 pKa = 11.84AVVADD252 pKa = 4.17SVVPPQMNLADD263 pKa = 4.23TLWPNAARR271 pKa = 11.84GFDD274 pKa = 3.82ALEE277 pKa = 4.43AACDD281 pKa = 3.83AQPACQAMLPDD292 pKa = 4.02LTGTLATTVTDD303 pKa = 4.93LDD305 pKa = 3.47AHH307 pKa = 5.86PQVVSVTAGDD317 pKa = 4.21GSPVDD322 pKa = 3.94VMVDD326 pKa = 3.78GYY328 pKa = 11.72KK329 pKa = 10.37LASLVNSASLATNGLVDD346 pKa = 4.15MPAIIAAAGSGDD358 pKa = 3.82VTPAAQLLGSAQPSTVPLVGSGLTYY383 pKa = 10.84GVFCGEE389 pKa = 4.14AVAHH393 pKa = 6.23TSAEE397 pKa = 4.09AMFGAAKK404 pKa = 10.43NSLPQFPDD412 pKa = 2.97AVLSLTPQLPWLVGDD427 pKa = 4.31CAAWNVDD434 pKa = 3.53AVPDD438 pKa = 4.69LVVQPANSTVPVLLLSGGLDD458 pKa = 3.35GVTPPGNADD467 pKa = 3.01IAKK470 pKa = 8.2ATLPNSVALTFPEE483 pKa = 4.55SGHH486 pKa = 6.39GVLTQSDD493 pKa = 4.44CGPAAVTGFLDD504 pKa = 5.04DD505 pKa = 4.38PSASYY510 pKa = 10.63RR511 pKa = 11.84PQCLDD516 pKa = 3.35TVVIPPFSTSLVGSGG531 pKa = 3.6
Molecular weight: 54.39 kDa
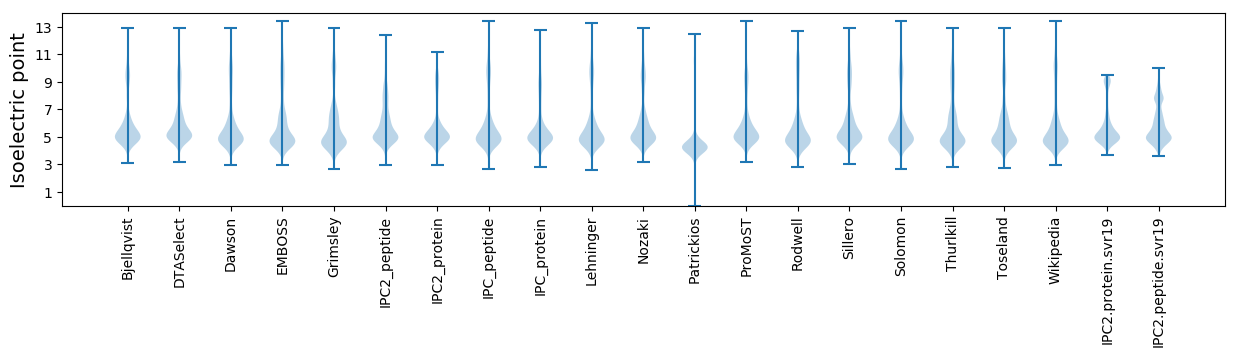
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A235G705|A0A235G705_9NOCA Amino acid/polyamine/organocation transporter (APC superfamily) OS=Rhodococcus sp. OK302 OX=1882769 GN=BDB13_2916 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
Molecular weight: 4.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1987623 |
29 |
9545 |
323.3 |
34.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.061 ± 0.039 | 0.783 ± 0.01 |
6.095 ± 0.028 | 5.458 ± 0.027 |
3.165 ± 0.019 | 8.806 ± 0.033 |
2.076 ± 0.016 | 4.816 ± 0.022 |
2.478 ± 0.025 | 9.763 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.015 | 2.509 ± 0.018 |
5.324 ± 0.022 | 2.883 ± 0.016 |
6.536 ± 0.035 | 6.474 ± 0.025 |
6.429 ± 0.039 | 8.765 ± 0.035 |
1.423 ± 0.011 | 2.067 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
