
Sidastrum golden leaf spot virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 9.24
Get precalculated fractions of proteins
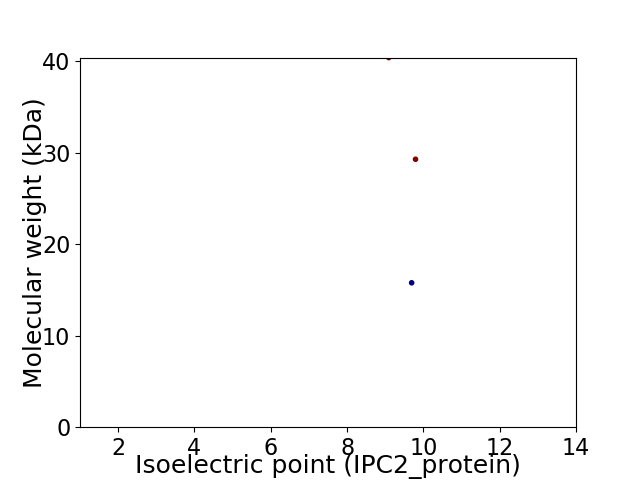
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9KZK5|E9KZK5_9GEMI Replication-associated protein OS=Sidastrum golden leaf spot virus OX=858513 GN=AC1 PE=4 SV=1
MM1 pKa = 7.74PSALFQRR8 pKa = 11.84KK9 pKa = 7.51KK10 pKa = 10.21HH11 pKa = 5.52FPNYY15 pKa = 10.03LPSKK19 pKa = 9.07PQLIKK24 pKa = 10.92NISEE28 pKa = 4.31YY29 pKa = 11.05VEE31 pKa = 5.27SSMKK35 pKa = 9.45MGNLIYY41 pKa = 9.98TSCFSSKK48 pKa = 10.33GSSHH52 pKa = 6.35VRR54 pKa = 11.84IADD57 pKa = 3.57SSILNIPAIPMYY69 pKa = 11.22VMEE72 pKa = 4.82NTNHH76 pKa = 6.91ASPPQTPRR84 pKa = 11.84HH85 pKa = 5.1ISRR88 pKa = 11.84RR89 pKa = 11.84TEE91 pKa = 3.21ITANGVVFRR100 pKa = 11.84STEE103 pKa = 3.78DD104 pKa = 3.47LLEE107 pKa = 4.92GGQQTDD113 pKa = 4.14DD114 pKa = 4.32DD115 pKa = 4.44ATTAPLNAALYY126 pKa = 10.49EE127 pKa = 4.1EE128 pKa = 4.7TLLKK132 pKa = 10.54SSKK135 pKa = 10.03PEE137 pKa = 3.27IRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84TLSSYY146 pKa = 11.39HH147 pKa = 6.25NIKK150 pKa = 10.94ANIEE154 pKa = 4.01RR155 pKa = 11.84LFQKK159 pKa = 10.5APEE162 pKa = 4.11PWTPPFQLSSFTRR175 pKa = 11.84VPDD178 pKa = 4.46EE179 pKa = 3.99MQEE182 pKa = 3.85WADD185 pKa = 3.78DD186 pKa = 3.86YY187 pKa = 11.3FGRR190 pKa = 11.84GAAARR195 pKa = 11.84PGKK198 pKa = 8.52TDD200 pKa = 2.96EE201 pKa = 3.78YY202 pKa = 10.18HH203 pKa = 6.07RR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84VIVGRR211 pKa = 11.84GRR213 pKa = 11.84QCGLVRR219 pKa = 11.84LAHH222 pKa = 6.69IIILADD228 pKa = 3.08TWTSILGFTQTKK240 pKa = 10.39RR241 pKa = 11.84NITSSMMSHH250 pKa = 6.76RR251 pKa = 11.84NILRR255 pKa = 11.84LSTGKK260 pKa = 9.9NLLGPKK266 pKa = 8.71KK267 pKa = 9.16TGSQIANTEE276 pKa = 4.15SQFKK280 pKa = 10.35LKK282 pKa = 10.48EE283 pKa = 4.19GSHH286 pKa = 5.47QSCSAIQARR295 pKa = 11.84GPAIKK300 pKa = 10.02ISLTKK305 pKa = 10.52RR306 pKa = 11.84KK307 pKa = 8.25MLRR310 pKa = 11.84LKK312 pKa = 10.46SWTLHH317 pKa = 4.46NAKK320 pKa = 10.29FIFLDD325 pKa = 3.49SPLYY329 pKa = 8.71QTSTQDD335 pKa = 3.61CEE337 pKa = 4.47KK338 pKa = 10.45EE339 pKa = 4.41SNSDD343 pKa = 2.94GDD345 pKa = 4.23GFTSSAAAPYY355 pKa = 9.65TSTSTAA361 pKa = 3.19
MM1 pKa = 7.74PSALFQRR8 pKa = 11.84KK9 pKa = 7.51KK10 pKa = 10.21HH11 pKa = 5.52FPNYY15 pKa = 10.03LPSKK19 pKa = 9.07PQLIKK24 pKa = 10.92NISEE28 pKa = 4.31YY29 pKa = 11.05VEE31 pKa = 5.27SSMKK35 pKa = 9.45MGNLIYY41 pKa = 9.98TSCFSSKK48 pKa = 10.33GSSHH52 pKa = 6.35VRR54 pKa = 11.84IADD57 pKa = 3.57SSILNIPAIPMYY69 pKa = 11.22VMEE72 pKa = 4.82NTNHH76 pKa = 6.91ASPPQTPRR84 pKa = 11.84HH85 pKa = 5.1ISRR88 pKa = 11.84RR89 pKa = 11.84TEE91 pKa = 3.21ITANGVVFRR100 pKa = 11.84STEE103 pKa = 3.78DD104 pKa = 3.47LLEE107 pKa = 4.92GGQQTDD113 pKa = 4.14DD114 pKa = 4.32DD115 pKa = 4.44ATTAPLNAALYY126 pKa = 10.49EE127 pKa = 4.1EE128 pKa = 4.7TLLKK132 pKa = 10.54SSKK135 pKa = 10.03PEE137 pKa = 3.27IRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84TLSSYY146 pKa = 11.39HH147 pKa = 6.25NIKK150 pKa = 10.94ANIEE154 pKa = 4.01RR155 pKa = 11.84LFQKK159 pKa = 10.5APEE162 pKa = 4.11PWTPPFQLSSFTRR175 pKa = 11.84VPDD178 pKa = 4.46EE179 pKa = 3.99MQEE182 pKa = 3.85WADD185 pKa = 3.78DD186 pKa = 3.86YY187 pKa = 11.3FGRR190 pKa = 11.84GAAARR195 pKa = 11.84PGKK198 pKa = 8.52TDD200 pKa = 2.96EE201 pKa = 3.78YY202 pKa = 10.18HH203 pKa = 6.07RR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84VIVGRR211 pKa = 11.84GRR213 pKa = 11.84QCGLVRR219 pKa = 11.84LAHH222 pKa = 6.69IIILADD228 pKa = 3.08TWTSILGFTQTKK240 pKa = 10.39RR241 pKa = 11.84NITSSMMSHH250 pKa = 6.76RR251 pKa = 11.84NILRR255 pKa = 11.84LSTGKK260 pKa = 9.9NLLGPKK266 pKa = 8.71KK267 pKa = 9.16TGSQIANTEE276 pKa = 4.15SQFKK280 pKa = 10.35LKK282 pKa = 10.48EE283 pKa = 4.19GSHH286 pKa = 5.47QSCSAIQARR295 pKa = 11.84GPAIKK300 pKa = 10.02ISLTKK305 pKa = 10.52RR306 pKa = 11.84KK307 pKa = 8.25MLRR310 pKa = 11.84LKK312 pKa = 10.46SWTLHH317 pKa = 4.46NAKK320 pKa = 10.29FIFLDD325 pKa = 3.49SPLYY329 pKa = 8.71QTSTQDD335 pKa = 3.61CEE337 pKa = 4.47KK338 pKa = 10.45EE339 pKa = 4.41SNSDD343 pKa = 2.94GDD345 pKa = 4.23GFTSSAAAPYY355 pKa = 9.65TSTSTAA361 pKa = 3.19
Molecular weight: 40.4 kDa
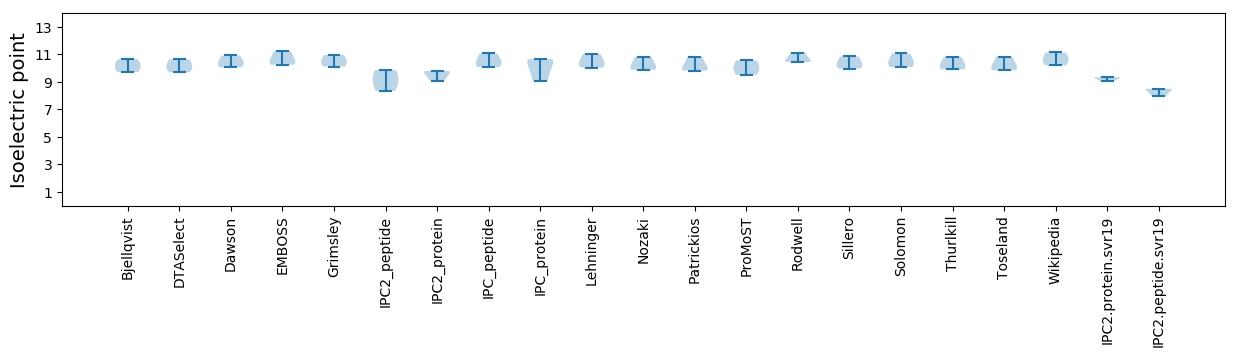
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2GI61|E2GI61_9GEMI Replication enhancer OS=Sidastrum golden leaf spot virus OX=858513 GN=AC3 PE=3 SV=2
MM1 pKa = 7.93IYY3 pKa = 10.96LKK5 pKa = 10.0MPKK8 pKa = 9.05RR9 pKa = 11.84DD10 pKa = 3.64PSWRR14 pKa = 11.84SMAGTSKK21 pKa = 10.59VSRR24 pKa = 11.84SSNFSPRR31 pKa = 11.84GGPKK35 pKa = 9.75VNKK38 pKa = 9.81ASEE41 pKa = 4.12WVNRR45 pKa = 11.84PMYY48 pKa = 9.79RR49 pKa = 11.84KK50 pKa = 8.95PRR52 pKa = 11.84IYY54 pKa = 9.6RR55 pKa = 11.84TLRR58 pKa = 11.84TPDD61 pKa = 3.21VPRR64 pKa = 11.84GCEE67 pKa = 4.34GPCKK71 pKa = 9.32VQSYY75 pKa = 6.98EE76 pKa = 3.66QRR78 pKa = 11.84NDD80 pKa = 2.52ISHH83 pKa = 6.96AGKK86 pKa = 10.32VICISDD92 pKa = 3.57VTRR95 pKa = 11.84GSGITHH101 pKa = 6.35RR102 pKa = 11.84VGKK105 pKa = 9.44RR106 pKa = 11.84FCVKK110 pKa = 9.92SVYY113 pKa = 10.16ILGKK117 pKa = 9.37IWMDD121 pKa = 3.39EE122 pKa = 4.0NIKK125 pKa = 10.61LKK127 pKa = 10.73NHH129 pKa = 5.95TNSVMFWLVRR139 pKa = 11.84DD140 pKa = 3.78RR141 pKa = 11.84RR142 pKa = 11.84PNGTSPWTLVICSTCSTMSLVLQLLRR168 pKa = 11.84MISEE172 pKa = 3.65IVIRR176 pKa = 11.84FNTDD180 pKa = 2.18SMPRR184 pKa = 11.84SLVVNMPAMSRR195 pKa = 11.84PWSGGFGRR203 pKa = 11.84SILMSCITTRR213 pKa = 11.84KK214 pKa = 10.01LLDD217 pKa = 3.08TRR219 pKa = 11.84TIRR222 pKa = 11.84RR223 pKa = 11.84MPSVVHH229 pKa = 5.56GHH231 pKa = 5.4VLMPLIPCMQLFKK244 pKa = 10.76IRR246 pKa = 11.84IYY248 pKa = 10.52FYY250 pKa = 11.18DD251 pKa = 4.08SISII255 pKa = 3.82
MM1 pKa = 7.93IYY3 pKa = 10.96LKK5 pKa = 10.0MPKK8 pKa = 9.05RR9 pKa = 11.84DD10 pKa = 3.64PSWRR14 pKa = 11.84SMAGTSKK21 pKa = 10.59VSRR24 pKa = 11.84SSNFSPRR31 pKa = 11.84GGPKK35 pKa = 9.75VNKK38 pKa = 9.81ASEE41 pKa = 4.12WVNRR45 pKa = 11.84PMYY48 pKa = 9.79RR49 pKa = 11.84KK50 pKa = 8.95PRR52 pKa = 11.84IYY54 pKa = 9.6RR55 pKa = 11.84TLRR58 pKa = 11.84TPDD61 pKa = 3.21VPRR64 pKa = 11.84GCEE67 pKa = 4.34GPCKK71 pKa = 9.32VQSYY75 pKa = 6.98EE76 pKa = 3.66QRR78 pKa = 11.84NDD80 pKa = 2.52ISHH83 pKa = 6.96AGKK86 pKa = 10.32VICISDD92 pKa = 3.57VTRR95 pKa = 11.84GSGITHH101 pKa = 6.35RR102 pKa = 11.84VGKK105 pKa = 9.44RR106 pKa = 11.84FCVKK110 pKa = 9.92SVYY113 pKa = 10.16ILGKK117 pKa = 9.37IWMDD121 pKa = 3.39EE122 pKa = 4.0NIKK125 pKa = 10.61LKK127 pKa = 10.73NHH129 pKa = 5.95TNSVMFWLVRR139 pKa = 11.84DD140 pKa = 3.78RR141 pKa = 11.84RR142 pKa = 11.84PNGTSPWTLVICSTCSTMSLVLQLLRR168 pKa = 11.84MISEE172 pKa = 3.65IVIRR176 pKa = 11.84FNTDD180 pKa = 2.18SMPRR184 pKa = 11.84SLVVNMPAMSRR195 pKa = 11.84PWSGGFGRR203 pKa = 11.84SILMSCITTRR213 pKa = 11.84KK214 pKa = 10.01LLDD217 pKa = 3.08TRR219 pKa = 11.84TIRR222 pKa = 11.84RR223 pKa = 11.84MPSVVHH229 pKa = 5.56GHH231 pKa = 5.4VLMPLIPCMQLFKK244 pKa = 10.76IRR246 pKa = 11.84IYY248 pKa = 10.52FYY250 pKa = 11.18DD251 pKa = 4.08SISII255 pKa = 3.82
Molecular weight: 29.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
749 |
133 |
361 |
249.7 |
28.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.073 ± 1.49 | 1.736 ± 0.622 |
3.471 ± 0.432 | 3.872 ± 0.811 |
3.738 ± 0.816 | 5.207 ± 0.524 |
3.204 ± 1.249 | 7.343 ± 0.416 |
5.607 ± 0.911 | 7.477 ± 0.354 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.139 ± 0.965 | 4.272 ± 0.155 |
5.207 ± 1.555 | 3.071 ± 0.797 |
8.678 ± 1.027 | 10.814 ± 0.63 |
7.477 ± 0.682 | 4.806 ± 1.396 |
1.736 ± 0.371 | 3.071 ± 0.499 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
