
Enterovirga rhinocerotis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Enterovirga
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
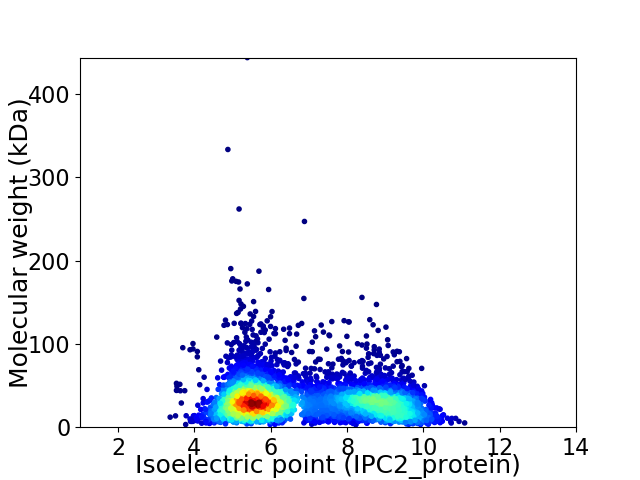
Virtual 2D-PAGE plot for 4822 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
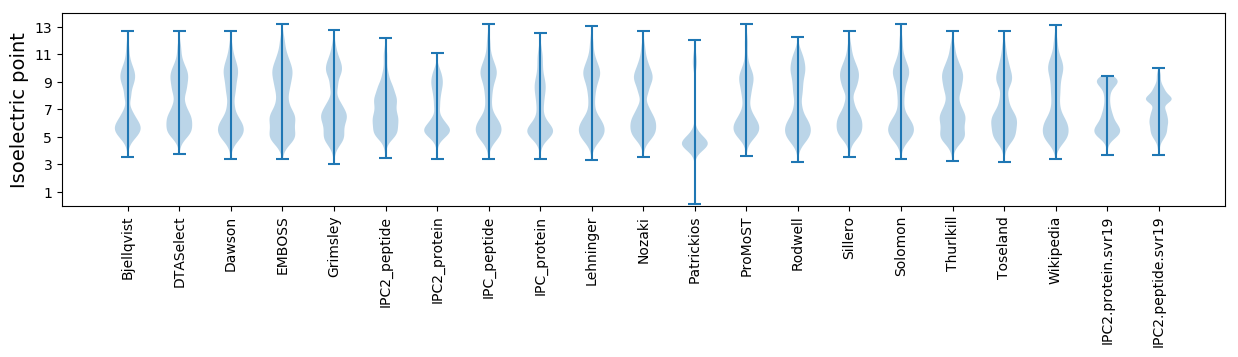
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R7C088|A0A4R7C088_9RHIZ Uncharacterized protein OS=Enterovirga rhinocerotis OX=1339210 GN=EV668_2723 PE=4 SV=1
MM1 pKa = 7.48AGVFRR6 pKa = 11.84FTSGGNYY13 pKa = 10.1DD14 pKa = 4.26PNNSSSIHH22 pKa = 6.34RR23 pKa = 11.84FNSGPPLDD31 pKa = 5.44LFINTNDD38 pKa = 3.46SMVVTASSYY47 pKa = 10.27FLAYY51 pKa = 10.46NSGFGTSGAWTLNAPDD67 pKa = 4.19GAYY70 pKa = 10.55LGPTQHH76 pKa = 7.42AIYY79 pKa = 10.32LDD81 pKa = 3.83YY82 pKa = 11.51SGGAGVANFQGGNWGDD98 pKa = 3.65VLIGGSAADD107 pKa = 3.85TLNGGGGDD115 pKa = 3.67DD116 pKa = 4.23RR117 pKa = 11.84LNGGLGADD125 pKa = 3.71TMTGGTGNDD134 pKa = 2.81IFYY137 pKa = 10.44VDD139 pKa = 3.83NVGDD143 pKa = 3.87IVVEE147 pKa = 3.9LAGQGTDD154 pKa = 3.48QVRR157 pKa = 11.84TTLDD161 pKa = 3.36NYY163 pKa = 10.67VLGANVEE170 pKa = 4.26NLFLMGTADD179 pKa = 5.18LSGTGNALNNVLQGNSGANTLSGGAGNDD207 pKa = 3.43RR208 pKa = 11.84LDD210 pKa = 3.56GRR212 pKa = 11.84AGADD216 pKa = 3.31TLIGGTGNDD225 pKa = 3.24TYY227 pKa = 11.51YY228 pKa = 10.22IDD230 pKa = 5.13DD231 pKa = 4.86LGDD234 pKa = 3.54VMVEE238 pKa = 4.18EE239 pKa = 4.89AGEE242 pKa = 4.66GYY244 pKa = 8.16DD245 pKa = 3.6TAFLSVDD252 pKa = 3.94GYY254 pKa = 11.59VLADD258 pKa = 3.66GASVEE263 pKa = 4.24RR264 pKa = 11.84LILRR268 pKa = 11.84DD269 pKa = 4.18GIVSASGNDD278 pKa = 3.69LDD280 pKa = 6.17NVLQGNAGDD289 pKa = 4.1NVLSGGAGRR298 pKa = 11.84DD299 pKa = 3.69TLKK302 pKa = 11.11GGAGNDD308 pKa = 3.91TLSGGAGIDD317 pKa = 3.44SLYY320 pKa = 10.98GGTGADD326 pKa = 3.51TFVLTATTADD336 pKa = 3.52RR337 pKa = 11.84DD338 pKa = 3.91MIRR341 pKa = 11.84DD342 pKa = 4.35FVAGEE347 pKa = 3.99DD348 pKa = 3.71TLQISASLFGGGLLAGDD365 pKa = 4.67ALTPEE370 pKa = 4.27QFVLSTTGRR379 pKa = 11.84ATSATDD385 pKa = 2.88RR386 pKa = 11.84FLYY389 pKa = 10.71VEE391 pKa = 4.63SKK393 pKa = 10.3GHH395 pKa = 7.08LYY397 pKa = 10.82YY398 pKa = 10.93DD399 pKa = 3.88ADD401 pKa = 3.68GSGRR405 pKa = 11.84DD406 pKa = 3.74EE407 pKa = 4.43APQLIAYY414 pKa = 7.51LTPNTALTASDD425 pKa = 4.46FYY427 pKa = 11.97VFF429 pKa = 4.82
MM1 pKa = 7.48AGVFRR6 pKa = 11.84FTSGGNYY13 pKa = 10.1DD14 pKa = 4.26PNNSSSIHH22 pKa = 6.34RR23 pKa = 11.84FNSGPPLDD31 pKa = 5.44LFINTNDD38 pKa = 3.46SMVVTASSYY47 pKa = 10.27FLAYY51 pKa = 10.46NSGFGTSGAWTLNAPDD67 pKa = 4.19GAYY70 pKa = 10.55LGPTQHH76 pKa = 7.42AIYY79 pKa = 10.32LDD81 pKa = 3.83YY82 pKa = 11.51SGGAGVANFQGGNWGDD98 pKa = 3.65VLIGGSAADD107 pKa = 3.85TLNGGGGDD115 pKa = 3.67DD116 pKa = 4.23RR117 pKa = 11.84LNGGLGADD125 pKa = 3.71TMTGGTGNDD134 pKa = 2.81IFYY137 pKa = 10.44VDD139 pKa = 3.83NVGDD143 pKa = 3.87IVVEE147 pKa = 3.9LAGQGTDD154 pKa = 3.48QVRR157 pKa = 11.84TTLDD161 pKa = 3.36NYY163 pKa = 10.67VLGANVEE170 pKa = 4.26NLFLMGTADD179 pKa = 5.18LSGTGNALNNVLQGNSGANTLSGGAGNDD207 pKa = 3.43RR208 pKa = 11.84LDD210 pKa = 3.56GRR212 pKa = 11.84AGADD216 pKa = 3.31TLIGGTGNDD225 pKa = 3.24TYY227 pKa = 11.51YY228 pKa = 10.22IDD230 pKa = 5.13DD231 pKa = 4.86LGDD234 pKa = 3.54VMVEE238 pKa = 4.18EE239 pKa = 4.89AGEE242 pKa = 4.66GYY244 pKa = 8.16DD245 pKa = 3.6TAFLSVDD252 pKa = 3.94GYY254 pKa = 11.59VLADD258 pKa = 3.66GASVEE263 pKa = 4.24RR264 pKa = 11.84LILRR268 pKa = 11.84DD269 pKa = 4.18GIVSASGNDD278 pKa = 3.69LDD280 pKa = 6.17NVLQGNAGDD289 pKa = 4.1NVLSGGAGRR298 pKa = 11.84DD299 pKa = 3.69TLKK302 pKa = 11.11GGAGNDD308 pKa = 3.91TLSGGAGIDD317 pKa = 3.44SLYY320 pKa = 10.98GGTGADD326 pKa = 3.51TFVLTATTADD336 pKa = 3.52RR337 pKa = 11.84DD338 pKa = 3.91MIRR341 pKa = 11.84DD342 pKa = 4.35FVAGEE347 pKa = 3.99DD348 pKa = 3.71TLQISASLFGGGLLAGDD365 pKa = 4.67ALTPEE370 pKa = 4.27QFVLSTTGRR379 pKa = 11.84ATSATDD385 pKa = 2.88RR386 pKa = 11.84FLYY389 pKa = 10.71VEE391 pKa = 4.63SKK393 pKa = 10.3GHH395 pKa = 7.08LYY397 pKa = 10.82YY398 pKa = 10.93DD399 pKa = 3.88ADD401 pKa = 3.68GSGRR405 pKa = 11.84DD406 pKa = 3.74EE407 pKa = 4.43APQLIAYY414 pKa = 7.51LTPNTALTASDD425 pKa = 4.46FYY427 pKa = 11.97VFF429 pKa = 4.82
Molecular weight: 43.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R7C693|A0A4R7C693_9RHIZ (2R 3S)-2-methylisocitrate dehydratase OS=Enterovirga rhinocerotis OX=1339210 GN=EV668_1367 PE=4 SV=1
MM1 pKa = 6.55VTLAKK6 pKa = 10.43SPARR10 pKa = 11.84SVSLSQGSAMGALAHH25 pKa = 6.47PRR27 pKa = 11.84APAQVRR33 pKa = 11.84AAPRR37 pKa = 11.84LPASLMRR44 pKa = 11.84MGLAGRR50 pKa = 11.84LAIAGVGTVLLWAAIGLGLAA70 pKa = 4.33
MM1 pKa = 6.55VTLAKK6 pKa = 10.43SPARR10 pKa = 11.84SVSLSQGSAMGALAHH25 pKa = 6.47PRR27 pKa = 11.84APAQVRR33 pKa = 11.84AAPRR37 pKa = 11.84LPASLMRR44 pKa = 11.84MGLAGRR50 pKa = 11.84LAIAGVGTVLLWAAIGLGLAA70 pKa = 4.33
Molecular weight: 6.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1527424 |
29 |
4184 |
316.8 |
34.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.495 ± 0.049 | 0.77 ± 0.01 |
5.565 ± 0.027 | 5.69 ± 0.033 |
3.542 ± 0.021 | 9.222 ± 0.037 |
1.878 ± 0.016 | 4.896 ± 0.027 |
2.735 ± 0.027 | 10.212 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.22 ± 0.014 | 2.086 ± 0.017 |
5.706 ± 0.032 | 2.631 ± 0.018 |
8.027 ± 0.039 | 5.301 ± 0.024 |
5.153 ± 0.022 | 7.525 ± 0.029 |
1.298 ± 0.013 | 2.049 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
