
Firmicutes bacterium CAG:56
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
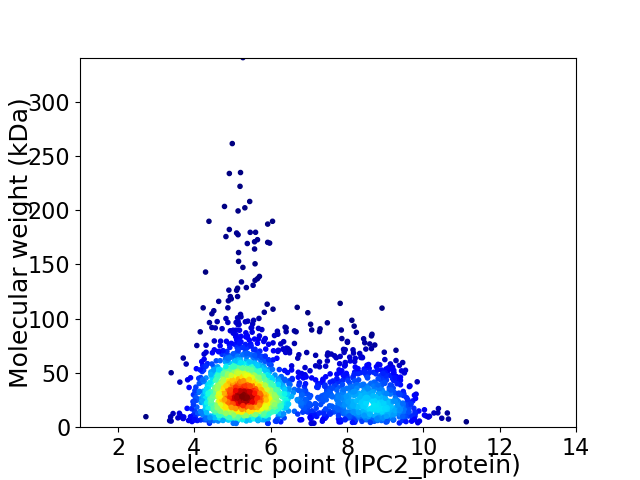
Virtual 2D-PAGE plot for 2160 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
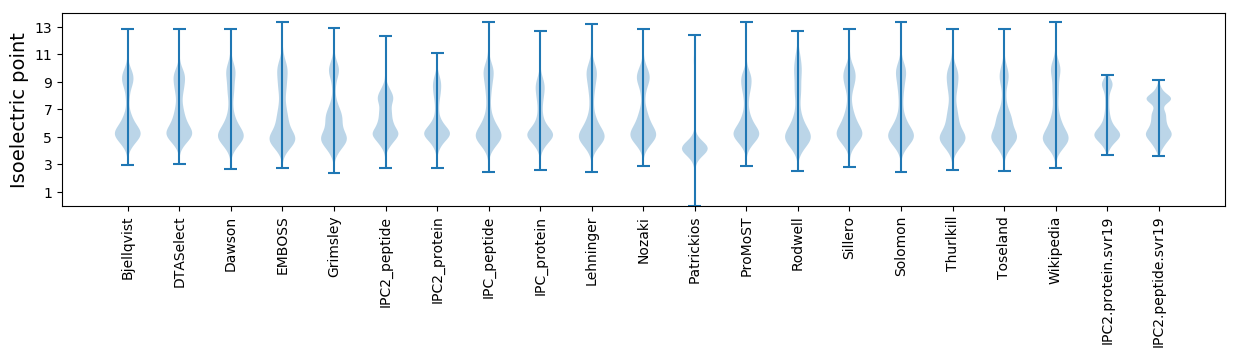
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6CGU4|R6CGU4_9FIRM Crispr-protein OS=Firmicutes bacterium CAG:56 OX=1263031 GN=BN708_01819 PE=4 SV=1
MM1 pKa = 7.26KK2 pKa = 10.15KK3 pKa = 10.17KK4 pKa = 10.71VIAGLLGTAMVASLLAGCGSSNNTTDD30 pKa = 3.12TTADD34 pKa = 3.12TTAATEE40 pKa = 3.98AAADD44 pKa = 3.93TTADD48 pKa = 3.53TTADD52 pKa = 3.27TAAADD57 pKa = 4.18AEE59 pKa = 4.61SEE61 pKa = 4.26AQSVADD67 pKa = 3.62AAGDD71 pKa = 3.66VDD73 pKa = 5.73LSNADD78 pKa = 3.6VAVMYY83 pKa = 7.24YY84 pKa = 9.96TYY86 pKa = 11.0GDD88 pKa = 3.58TYY90 pKa = 10.05IASVRR95 pKa = 11.84TALDD99 pKa = 3.21AKK101 pKa = 10.67LDD103 pKa = 3.95EE104 pKa = 5.56LGVKK108 pKa = 8.92YY109 pKa = 10.45QDD111 pKa = 3.17YY112 pKa = 10.55DD113 pKa = 3.77ANNNQTTQNEE123 pKa = 4.42QIDD126 pKa = 4.0TAIQTGANVLIVNIVTSGSVDD147 pKa = 2.99ASSQIVEE154 pKa = 4.21KK155 pKa = 10.89AKK157 pKa = 10.74AADD160 pKa = 3.24IPVIFFNRR168 pKa = 11.84AVEE171 pKa = 4.04GDD173 pKa = 3.51EE174 pKa = 5.93DD175 pKa = 3.74EE176 pKa = 5.2GKK178 pKa = 10.76VLGSYY183 pKa = 9.41DD184 pKa = 2.79KK185 pKa = 11.15CAFVGTDD192 pKa = 3.25APEE195 pKa = 4.49AGHH198 pKa = 5.95MQGEE202 pKa = 4.53MIGNYY207 pKa = 10.2LVDD210 pKa = 4.22NYY212 pKa = 11.46DD213 pKa = 3.74AVDD216 pKa = 3.9LNGDD220 pKa = 3.79GKK222 pKa = 10.56ISYY225 pKa = 10.98AMFMGQLGNVEE236 pKa = 4.61AIYY239 pKa = 8.58RR240 pKa = 11.84TQYY243 pKa = 10.72GVEE246 pKa = 4.18DD247 pKa = 3.83ANKK250 pKa = 9.65VLEE253 pKa = 4.5EE254 pKa = 3.99NGKK257 pKa = 9.65AALEE261 pKa = 4.23YY262 pKa = 10.29FDD264 pKa = 6.02SSNTDD269 pKa = 3.6CYY271 pKa = 11.0QVDD274 pKa = 3.69QDD276 pKa = 4.72GNWSATAANNYY287 pKa = 5.86MTTNLTQYY295 pKa = 11.04NEE297 pKa = 4.08EE298 pKa = 4.08NKK300 pKa = 10.64NMIEE304 pKa = 5.38LVICNNDD311 pKa = 2.77GMAEE315 pKa = 4.13GAISALNDD323 pKa = 3.01KK324 pKa = 10.44GYY326 pKa = 10.98NLGTDD331 pKa = 3.56DD332 pKa = 5.15CKK334 pKa = 10.52TIPVFGVDD342 pKa = 3.08ATDD345 pKa = 3.91AAKK348 pKa = 10.46QLIKK352 pKa = 10.31DD353 pKa = 3.79GKK355 pKa = 7.36MTGTIKK361 pKa = 10.46QDD363 pKa = 3.31AEE365 pKa = 4.21GMADD369 pKa = 4.88CIADD373 pKa = 3.65LTKK376 pKa = 10.89NAGSGQDD383 pKa = 3.33VMANTDD389 pKa = 3.34SYY391 pKa = 11.69NISEE395 pKa = 4.19NVKK398 pKa = 8.84NKK400 pKa = 10.15IYY402 pKa = 10.29IPYY405 pKa = 10.75AMYY408 pKa = 9.87TGEE411 pKa = 4.05EE412 pKa = 4.06
MM1 pKa = 7.26KK2 pKa = 10.15KK3 pKa = 10.17KK4 pKa = 10.71VIAGLLGTAMVASLLAGCGSSNNTTDD30 pKa = 3.12TTADD34 pKa = 3.12TTAATEE40 pKa = 3.98AAADD44 pKa = 3.93TTADD48 pKa = 3.53TTADD52 pKa = 3.27TAAADD57 pKa = 4.18AEE59 pKa = 4.61SEE61 pKa = 4.26AQSVADD67 pKa = 3.62AAGDD71 pKa = 3.66VDD73 pKa = 5.73LSNADD78 pKa = 3.6VAVMYY83 pKa = 7.24YY84 pKa = 9.96TYY86 pKa = 11.0GDD88 pKa = 3.58TYY90 pKa = 10.05IASVRR95 pKa = 11.84TALDD99 pKa = 3.21AKK101 pKa = 10.67LDD103 pKa = 3.95EE104 pKa = 5.56LGVKK108 pKa = 8.92YY109 pKa = 10.45QDD111 pKa = 3.17YY112 pKa = 10.55DD113 pKa = 3.77ANNNQTTQNEE123 pKa = 4.42QIDD126 pKa = 4.0TAIQTGANVLIVNIVTSGSVDD147 pKa = 2.99ASSQIVEE154 pKa = 4.21KK155 pKa = 10.89AKK157 pKa = 10.74AADD160 pKa = 3.24IPVIFFNRR168 pKa = 11.84AVEE171 pKa = 4.04GDD173 pKa = 3.51EE174 pKa = 5.93DD175 pKa = 3.74EE176 pKa = 5.2GKK178 pKa = 10.76VLGSYY183 pKa = 9.41DD184 pKa = 2.79KK185 pKa = 11.15CAFVGTDD192 pKa = 3.25APEE195 pKa = 4.49AGHH198 pKa = 5.95MQGEE202 pKa = 4.53MIGNYY207 pKa = 10.2LVDD210 pKa = 4.22NYY212 pKa = 11.46DD213 pKa = 3.74AVDD216 pKa = 3.9LNGDD220 pKa = 3.79GKK222 pKa = 10.56ISYY225 pKa = 10.98AMFMGQLGNVEE236 pKa = 4.61AIYY239 pKa = 8.58RR240 pKa = 11.84TQYY243 pKa = 10.72GVEE246 pKa = 4.18DD247 pKa = 3.83ANKK250 pKa = 9.65VLEE253 pKa = 4.5EE254 pKa = 3.99NGKK257 pKa = 9.65AALEE261 pKa = 4.23YY262 pKa = 10.29FDD264 pKa = 6.02SSNTDD269 pKa = 3.6CYY271 pKa = 11.0QVDD274 pKa = 3.69QDD276 pKa = 4.72GNWSATAANNYY287 pKa = 5.86MTTNLTQYY295 pKa = 11.04NEE297 pKa = 4.08EE298 pKa = 4.08NKK300 pKa = 10.64NMIEE304 pKa = 5.38LVICNNDD311 pKa = 2.77GMAEE315 pKa = 4.13GAISALNDD323 pKa = 3.01KK324 pKa = 10.44GYY326 pKa = 10.98NLGTDD331 pKa = 3.56DD332 pKa = 5.15CKK334 pKa = 10.52TIPVFGVDD342 pKa = 3.08ATDD345 pKa = 3.91AAKK348 pKa = 10.46QLIKK352 pKa = 10.31DD353 pKa = 3.79GKK355 pKa = 7.36MTGTIKK361 pKa = 10.46QDD363 pKa = 3.31AEE365 pKa = 4.21GMADD369 pKa = 4.88CIADD373 pKa = 3.65LTKK376 pKa = 10.89NAGSGQDD383 pKa = 3.33VMANTDD389 pKa = 3.34SYY391 pKa = 11.69NISEE395 pKa = 4.19NVKK398 pKa = 8.84NKK400 pKa = 10.15IYY402 pKa = 10.29IPYY405 pKa = 10.75AMYY408 pKa = 9.87TGEE411 pKa = 4.05EE412 pKa = 4.06
Molecular weight: 43.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6CHB6|R6CHB6_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:56 OX=1263031 GN=BN708_01967 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
707107 |
29 |
3033 |
327.4 |
36.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.882 ± 0.062 | 1.512 ± 0.027 |
5.715 ± 0.046 | 7.701 ± 0.067 |
3.907 ± 0.04 | 7.052 ± 0.039 |
1.707 ± 0.026 | 6.972 ± 0.056 |
7.579 ± 0.058 | 8.523 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.023 ± 0.035 | 4.219 ± 0.043 |
3.089 ± 0.028 | 3.487 ± 0.027 |
4.261 ± 0.053 | 5.753 ± 0.055 |
5.718 ± 0.069 | 6.923 ± 0.043 |
0.871 ± 0.017 | 4.092 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
