
Actinomycetales bacterium JB111
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; unclassified Actinomycetales
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
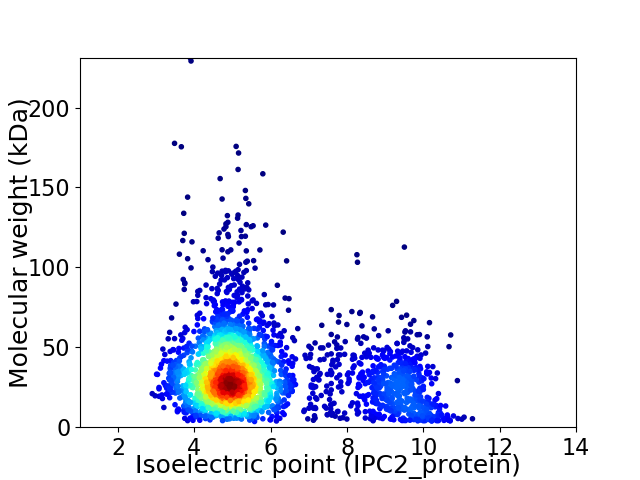
Virtual 2D-PAGE plot for 2923 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1R4F4C1|A0A1R4F4C1_9ACTO 30S ribosomal protein S4 OS=Actinomycetales bacterium JB111 OX=1434822 GN=rpsD PE=3 SV=1
MM1 pKa = 7.63AGADD5 pKa = 4.26RR6 pKa = 11.84LPLPDD11 pKa = 5.86GIATTLLVQGTSISRR26 pKa = 11.84IQGRR30 pKa = 11.84LEE32 pKa = 3.67QTLLSDD38 pKa = 3.97GSASQRR44 pKa = 11.84LRR46 pKa = 11.84GAFPAGLSAEE56 pKa = 4.77AIDD59 pKa = 4.32TEE61 pKa = 4.48LSVLSGHH68 pKa = 6.42VSTATSNMGSAGQHH82 pKa = 4.56VWDD85 pKa = 4.32VGAALEE91 pKa = 4.39TCEE94 pKa = 5.04SEE96 pKa = 5.17VEE98 pKa = 4.01GLQADD103 pKa = 4.09WDD105 pKa = 4.1EE106 pKa = 4.33LDD108 pKa = 3.25AWFDD112 pKa = 3.59RR113 pKa = 11.84ALADD117 pKa = 3.81INSDD121 pKa = 3.29PYY123 pKa = 10.77YY124 pKa = 10.86AEE126 pKa = 4.98PEE128 pKa = 4.18NQRR131 pKa = 11.84DD132 pKa = 3.26RR133 pKa = 11.84AEE135 pKa = 4.59AIEE138 pKa = 5.06SIDD141 pKa = 3.65LQRR144 pKa = 11.84EE145 pKa = 4.03SGRR148 pKa = 11.84DD149 pKa = 3.54DD150 pKa = 3.74LKK152 pKa = 11.16EE153 pKa = 4.22RR154 pKa = 11.84YY155 pKa = 8.96HH156 pKa = 6.99SAIDD160 pKa = 3.68TFGDD164 pKa = 3.57AASAAATRR172 pKa = 11.84LTALTDD178 pKa = 4.72GIVPPSQQGSVTDD191 pKa = 3.73IAAAMTAGTTIADD204 pKa = 3.87HH205 pKa = 7.57DD206 pKa = 4.42RR207 pKa = 11.84ATHH210 pKa = 4.65VAEE213 pKa = 4.58EE214 pKa = 4.07QAEE217 pKa = 4.23DD218 pKa = 4.25ASDD221 pKa = 3.64LLEE224 pKa = 4.42RR225 pKa = 11.84ADD227 pKa = 5.15DD228 pKa = 3.93EE229 pKa = 4.63SLSAEE234 pKa = 4.15EE235 pKa = 3.83RR236 pKa = 11.84QEE238 pKa = 5.4AIDD241 pKa = 4.46EE242 pKa = 4.42FNATYY247 pKa = 11.02GDD249 pKa = 4.24LADD252 pKa = 5.08DD253 pKa = 5.06PYY255 pKa = 11.43FSAALMDD262 pKa = 4.77EE263 pKa = 4.28IGEE266 pKa = 4.26EE267 pKa = 4.0EE268 pKa = 4.58LMQHH272 pKa = 6.63IANLQDD278 pKa = 3.64YY279 pKa = 10.43QGEE282 pKa = 4.34NVEE285 pKa = 4.39NEE287 pKa = 4.35SVDD290 pKa = 3.85ASIDD294 pKa = 3.37TYY296 pKa = 11.76LNVLGSAFVTSTNDD310 pKa = 3.3PVGLSDD316 pKa = 3.87YY317 pKa = 9.87ATEE320 pKa = 4.33RR321 pKa = 11.84LTPAAEE327 pKa = 3.96TRR329 pKa = 11.84RR330 pKa = 11.84DD331 pKa = 3.74EE332 pKa = 4.22FTAAADD338 pKa = 3.65EE339 pKa = 4.52YY340 pKa = 11.52YY341 pKa = 10.06PYY343 pKa = 10.85DD344 pKa = 4.69DD345 pKa = 5.34GNGHH349 pKa = 5.96VPGWWSIGQLINGAQFADD367 pKa = 3.98VPVTPDD373 pKa = 2.95RR374 pKa = 11.84AFTEE378 pKa = 4.3SLGADD383 pKa = 3.3MMRR386 pKa = 11.84FYY388 pKa = 11.16YY389 pKa = 10.73DD390 pKa = 3.51PDD392 pKa = 3.99TVVGMQTNNPMNMSPTGNPYY412 pKa = 10.36PYY414 pKa = 10.18DD415 pKa = 3.58SLVEE419 pKa = 4.29FPEE422 pKa = 4.52GLEE425 pKa = 4.1LPGGVDD431 pKa = 3.52PEE433 pKa = 4.39QMFRR437 pKa = 11.84DD438 pKa = 3.84PLYY441 pKa = 10.38SLNYY445 pKa = 9.94SVTGDD450 pKa = 3.44EE451 pKa = 4.16EE452 pKa = 4.52AATGLYY458 pKa = 10.06LSEE461 pKa = 4.55VGTHH465 pKa = 5.46EE466 pKa = 4.35VNGEE470 pKa = 4.01QVPVSGMEE478 pKa = 4.12YY479 pKa = 10.39LMHH482 pKa = 6.93RR483 pKa = 11.84AVNYY487 pKa = 9.22EE488 pKa = 3.97PAYY491 pKa = 10.09HH492 pKa = 6.74EE493 pKa = 5.23AMLSPVVEE501 pKa = 4.6GAHH504 pKa = 5.42NVARR508 pKa = 11.84DD509 pKa = 3.76PYY511 pKa = 10.53DD512 pKa = 3.61EE513 pKa = 5.51EE514 pKa = 4.54STQVAQEE521 pKa = 3.94FVDD524 pKa = 3.21AHH526 pKa = 6.26LRR528 pKa = 11.84LIGGEE533 pKa = 4.01YY534 pKa = 10.14EE535 pKa = 4.38NYY537 pKa = 10.35VDD539 pKa = 3.81TTFMTTSVADD549 pKa = 3.29VWAANIDD556 pKa = 3.8SLMYY560 pKa = 10.76AVDD563 pKa = 4.22DD564 pKa = 4.9DD565 pKa = 4.45GVAVGEE571 pKa = 4.34DD572 pKa = 3.32GVYY575 pKa = 10.61AGNGPTQADD584 pKa = 3.99YY585 pKa = 10.38MFSISDD591 pKa = 3.59DD592 pKa = 3.33VRR594 pKa = 11.84AYY596 pKa = 10.98LPDD599 pKa = 3.36LMNYY603 pKa = 9.2FAEE606 pKa = 4.42DD607 pKa = 3.51RR608 pKa = 11.84PGEE611 pKa = 4.06FVIDD615 pKa = 4.18PNGPDD620 pKa = 3.58AGNPPALQRR629 pKa = 11.84IIDD632 pKa = 3.78ATVLDD637 pKa = 4.19SHH639 pKa = 7.47FDD641 pKa = 3.3AQNIFDD647 pKa = 4.35PEE649 pKa = 4.63HH650 pKa = 5.65YY651 pKa = 9.94QSLSGAEE658 pKa = 3.9NALQVSTDD666 pKa = 3.44QSSLFLNYY674 pKa = 9.97VNEE677 pKa = 4.28SLDD680 pKa = 3.54QQGAADD686 pKa = 4.43DD687 pKa = 4.35AQLEE691 pKa = 4.25QTRR694 pKa = 11.84GYY696 pKa = 10.56INGAVSGASGLIAGLGTTGRR716 pKa = 11.84FVYY719 pKa = 10.4GMADD723 pKa = 4.1DD724 pKa = 4.97LVMDD728 pKa = 5.14AFWDD732 pKa = 4.04AVGEE736 pKa = 4.23NYY738 pKa = 10.76DD739 pKa = 3.88PANASNSAVNFNSALEE755 pKa = 4.08DD756 pKa = 3.16AVYY759 pKa = 10.4QGAYY763 pKa = 9.46SAEE766 pKa = 4.17PWSVDD771 pKa = 2.67PWDD774 pKa = 4.22AQTNPAGVMSPEE786 pKa = 3.93QWSEE790 pKa = 3.59ATGQPRR796 pKa = 11.84FDD798 pKa = 4.1TDD800 pKa = 4.51NPADD804 pKa = 3.4WTPNQRR810 pKa = 11.84RR811 pKa = 11.84AYY813 pKa = 8.24EE814 pKa = 4.38AYY816 pKa = 10.26AIDD819 pKa = 4.33PNGGMGNYY827 pKa = 9.0TSGPVTEE834 pKa = 5.21IGDD837 pKa = 3.88SATEE841 pKa = 4.01AEE843 pKa = 4.59QLVTAHH849 pKa = 7.69RR850 pKa = 11.84SQADD854 pKa = 3.22QQ855 pKa = 3.31
MM1 pKa = 7.63AGADD5 pKa = 4.26RR6 pKa = 11.84LPLPDD11 pKa = 5.86GIATTLLVQGTSISRR26 pKa = 11.84IQGRR30 pKa = 11.84LEE32 pKa = 3.67QTLLSDD38 pKa = 3.97GSASQRR44 pKa = 11.84LRR46 pKa = 11.84GAFPAGLSAEE56 pKa = 4.77AIDD59 pKa = 4.32TEE61 pKa = 4.48LSVLSGHH68 pKa = 6.42VSTATSNMGSAGQHH82 pKa = 4.56VWDD85 pKa = 4.32VGAALEE91 pKa = 4.39TCEE94 pKa = 5.04SEE96 pKa = 5.17VEE98 pKa = 4.01GLQADD103 pKa = 4.09WDD105 pKa = 4.1EE106 pKa = 4.33LDD108 pKa = 3.25AWFDD112 pKa = 3.59RR113 pKa = 11.84ALADD117 pKa = 3.81INSDD121 pKa = 3.29PYY123 pKa = 10.77YY124 pKa = 10.86AEE126 pKa = 4.98PEE128 pKa = 4.18NQRR131 pKa = 11.84DD132 pKa = 3.26RR133 pKa = 11.84AEE135 pKa = 4.59AIEE138 pKa = 5.06SIDD141 pKa = 3.65LQRR144 pKa = 11.84EE145 pKa = 4.03SGRR148 pKa = 11.84DD149 pKa = 3.54DD150 pKa = 3.74LKK152 pKa = 11.16EE153 pKa = 4.22RR154 pKa = 11.84YY155 pKa = 8.96HH156 pKa = 6.99SAIDD160 pKa = 3.68TFGDD164 pKa = 3.57AASAAATRR172 pKa = 11.84LTALTDD178 pKa = 4.72GIVPPSQQGSVTDD191 pKa = 3.73IAAAMTAGTTIADD204 pKa = 3.87HH205 pKa = 7.57DD206 pKa = 4.42RR207 pKa = 11.84ATHH210 pKa = 4.65VAEE213 pKa = 4.58EE214 pKa = 4.07QAEE217 pKa = 4.23DD218 pKa = 4.25ASDD221 pKa = 3.64LLEE224 pKa = 4.42RR225 pKa = 11.84ADD227 pKa = 5.15DD228 pKa = 3.93EE229 pKa = 4.63SLSAEE234 pKa = 4.15EE235 pKa = 3.83RR236 pKa = 11.84QEE238 pKa = 5.4AIDD241 pKa = 4.46EE242 pKa = 4.42FNATYY247 pKa = 11.02GDD249 pKa = 4.24LADD252 pKa = 5.08DD253 pKa = 5.06PYY255 pKa = 11.43FSAALMDD262 pKa = 4.77EE263 pKa = 4.28IGEE266 pKa = 4.26EE267 pKa = 4.0EE268 pKa = 4.58LMQHH272 pKa = 6.63IANLQDD278 pKa = 3.64YY279 pKa = 10.43QGEE282 pKa = 4.34NVEE285 pKa = 4.39NEE287 pKa = 4.35SVDD290 pKa = 3.85ASIDD294 pKa = 3.37TYY296 pKa = 11.76LNVLGSAFVTSTNDD310 pKa = 3.3PVGLSDD316 pKa = 3.87YY317 pKa = 9.87ATEE320 pKa = 4.33RR321 pKa = 11.84LTPAAEE327 pKa = 3.96TRR329 pKa = 11.84RR330 pKa = 11.84DD331 pKa = 3.74EE332 pKa = 4.22FTAAADD338 pKa = 3.65EE339 pKa = 4.52YY340 pKa = 11.52YY341 pKa = 10.06PYY343 pKa = 10.85DD344 pKa = 4.69DD345 pKa = 5.34GNGHH349 pKa = 5.96VPGWWSIGQLINGAQFADD367 pKa = 3.98VPVTPDD373 pKa = 2.95RR374 pKa = 11.84AFTEE378 pKa = 4.3SLGADD383 pKa = 3.3MMRR386 pKa = 11.84FYY388 pKa = 11.16YY389 pKa = 10.73DD390 pKa = 3.51PDD392 pKa = 3.99TVVGMQTNNPMNMSPTGNPYY412 pKa = 10.36PYY414 pKa = 10.18DD415 pKa = 3.58SLVEE419 pKa = 4.29FPEE422 pKa = 4.52GLEE425 pKa = 4.1LPGGVDD431 pKa = 3.52PEE433 pKa = 4.39QMFRR437 pKa = 11.84DD438 pKa = 3.84PLYY441 pKa = 10.38SLNYY445 pKa = 9.94SVTGDD450 pKa = 3.44EE451 pKa = 4.16EE452 pKa = 4.52AATGLYY458 pKa = 10.06LSEE461 pKa = 4.55VGTHH465 pKa = 5.46EE466 pKa = 4.35VNGEE470 pKa = 4.01QVPVSGMEE478 pKa = 4.12YY479 pKa = 10.39LMHH482 pKa = 6.93RR483 pKa = 11.84AVNYY487 pKa = 9.22EE488 pKa = 3.97PAYY491 pKa = 10.09HH492 pKa = 6.74EE493 pKa = 5.23AMLSPVVEE501 pKa = 4.6GAHH504 pKa = 5.42NVARR508 pKa = 11.84DD509 pKa = 3.76PYY511 pKa = 10.53DD512 pKa = 3.61EE513 pKa = 5.51EE514 pKa = 4.54STQVAQEE521 pKa = 3.94FVDD524 pKa = 3.21AHH526 pKa = 6.26LRR528 pKa = 11.84LIGGEE533 pKa = 4.01YY534 pKa = 10.14EE535 pKa = 4.38NYY537 pKa = 10.35VDD539 pKa = 3.81TTFMTTSVADD549 pKa = 3.29VWAANIDD556 pKa = 3.8SLMYY560 pKa = 10.76AVDD563 pKa = 4.22DD564 pKa = 4.9DD565 pKa = 4.45GVAVGEE571 pKa = 4.34DD572 pKa = 3.32GVYY575 pKa = 10.61AGNGPTQADD584 pKa = 3.99YY585 pKa = 10.38MFSISDD591 pKa = 3.59DD592 pKa = 3.33VRR594 pKa = 11.84AYY596 pKa = 10.98LPDD599 pKa = 3.36LMNYY603 pKa = 9.2FAEE606 pKa = 4.42DD607 pKa = 3.51RR608 pKa = 11.84PGEE611 pKa = 4.06FVIDD615 pKa = 4.18PNGPDD620 pKa = 3.58AGNPPALQRR629 pKa = 11.84IIDD632 pKa = 3.78ATVLDD637 pKa = 4.19SHH639 pKa = 7.47FDD641 pKa = 3.3AQNIFDD647 pKa = 4.35PEE649 pKa = 4.63HH650 pKa = 5.65YY651 pKa = 9.94QSLSGAEE658 pKa = 3.9NALQVSTDD666 pKa = 3.44QSSLFLNYY674 pKa = 9.97VNEE677 pKa = 4.28SLDD680 pKa = 3.54QQGAADD686 pKa = 4.43DD687 pKa = 4.35AQLEE691 pKa = 4.25QTRR694 pKa = 11.84GYY696 pKa = 10.56INGAVSGASGLIAGLGTTGRR716 pKa = 11.84FVYY719 pKa = 10.4GMADD723 pKa = 4.1DD724 pKa = 4.97LVMDD728 pKa = 5.14AFWDD732 pKa = 4.04AVGEE736 pKa = 4.23NYY738 pKa = 10.76DD739 pKa = 3.88PANASNSAVNFNSALEE755 pKa = 4.08DD756 pKa = 3.16AVYY759 pKa = 10.4QGAYY763 pKa = 9.46SAEE766 pKa = 4.17PWSVDD771 pKa = 2.67PWDD774 pKa = 4.22AQTNPAGVMSPEE786 pKa = 3.93QWSEE790 pKa = 3.59ATGQPRR796 pKa = 11.84FDD798 pKa = 4.1TDD800 pKa = 4.51NPADD804 pKa = 3.4WTPNQRR810 pKa = 11.84RR811 pKa = 11.84AYY813 pKa = 8.24EE814 pKa = 4.38AYY816 pKa = 10.26AIDD819 pKa = 4.33PNGGMGNYY827 pKa = 9.0TSGPVTEE834 pKa = 5.21IGDD837 pKa = 3.88SATEE841 pKa = 4.01AEE843 pKa = 4.59QLVTAHH849 pKa = 7.69RR850 pKa = 11.84SQADD854 pKa = 3.22QQ855 pKa = 3.31
Molecular weight: 92.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1R4F9K3|A0A1R4F9K3_9ACTO Ribosomal RNA small subunit methyltransferase G OS=Actinomycetales bacterium JB111 OX=1434822 GN=rsmG PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.64GRR40 pKa = 11.84AKK42 pKa = 10.67LSAA45 pKa = 3.92
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.64GRR40 pKa = 11.84AKK42 pKa = 10.67LSAA45 pKa = 3.92
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
961640 |
37 |
2177 |
329.0 |
34.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.554 ± 0.076 | 0.504 ± 0.01 |
6.759 ± 0.046 | 6.038 ± 0.045 |
2.587 ± 0.027 | 9.715 ± 0.047 |
2.034 ± 0.021 | 3.966 ± 0.033 |
1.49 ± 0.031 | 9.767 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.826 ± 0.02 | 1.721 ± 0.024 |
5.721 ± 0.039 | 2.494 ± 0.023 |
7.531 ± 0.05 | 5.673 ± 0.032 |
6.241 ± 0.036 | 9.198 ± 0.046 |
1.382 ± 0.019 | 1.798 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
