
Hubei picorna-like virus 77
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
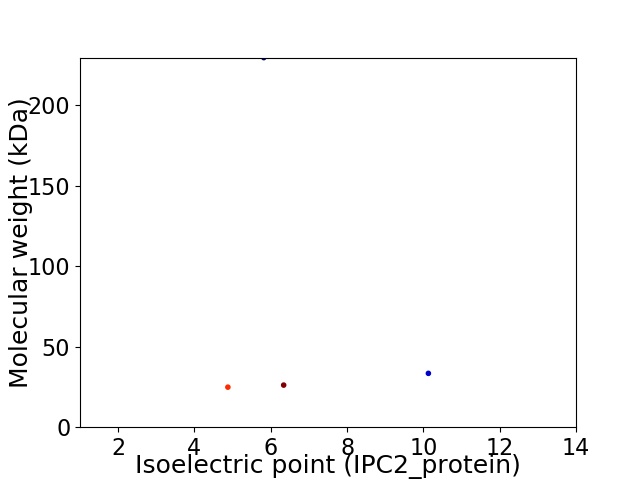
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
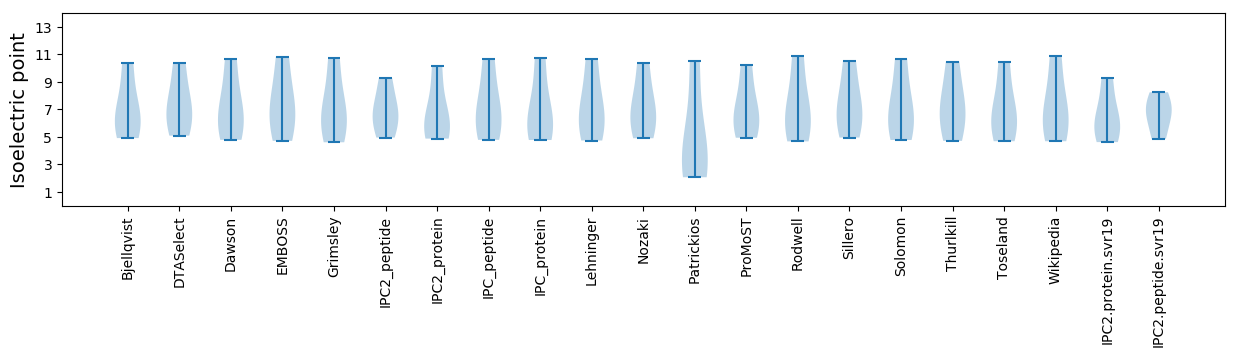
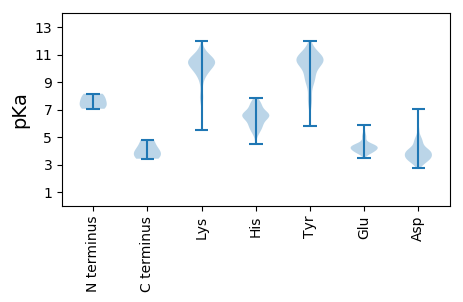
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KM48|A0A1L3KM48_9VIRU Calici_coat domain-containing protein OS=Hubei picorna-like virus 77 OX=1923161 PE=4 SV=1
MM1 pKa = 7.31SHH3 pKa = 7.51PGMSAAPAVSGDD15 pKa = 3.76PAISASTDD23 pKa = 3.16AAQLPLSTISTAPAPAAEE41 pKa = 5.44AIPHH45 pKa = 5.9TGSLNALDD53 pKa = 5.01PYY55 pKa = 11.03VKK57 pKa = 10.27EE58 pKa = 3.97QFISQGSFEE67 pKa = 4.65WTTADD72 pKa = 4.83LVGKK76 pKa = 9.95LIFSCPIHH84 pKa = 6.43PVNANPILAYY94 pKa = 10.28LSKK97 pKa = 10.38IYY99 pKa = 8.84NTWAGGLDD107 pKa = 4.3FKK109 pKa = 11.63AKK111 pKa = 9.55IAGTGFHH118 pKa = 7.01AGALALVRR126 pKa = 11.84LPPNVDD132 pKa = 3.14PATVSDD138 pKa = 4.13VNKK141 pKa = 7.92FTCFEE146 pKa = 4.22YY147 pKa = 10.73EE148 pKa = 4.06IIDD151 pKa = 4.06PKK153 pKa = 11.13LLEE156 pKa = 4.4CVSRR160 pKa = 11.84SVMDD164 pKa = 3.45QRR166 pKa = 11.84NTMYY170 pKa = 10.36HH171 pKa = 5.62YY172 pKa = 11.06NPYY175 pKa = 10.68DD176 pKa = 3.32ATNYY180 pKa = 10.63NSFGGNFCIFVLQALNTSSSGSTSISVQIFSKK212 pKa = 10.59ASEE215 pKa = 4.44DD216 pKa = 3.97FEE218 pKa = 4.38MLQIIPPNITTLSSTT233 pKa = 3.75
MM1 pKa = 7.31SHH3 pKa = 7.51PGMSAAPAVSGDD15 pKa = 3.76PAISASTDD23 pKa = 3.16AAQLPLSTISTAPAPAAEE41 pKa = 5.44AIPHH45 pKa = 5.9TGSLNALDD53 pKa = 5.01PYY55 pKa = 11.03VKK57 pKa = 10.27EE58 pKa = 3.97QFISQGSFEE67 pKa = 4.65WTTADD72 pKa = 4.83LVGKK76 pKa = 9.95LIFSCPIHH84 pKa = 6.43PVNANPILAYY94 pKa = 10.28LSKK97 pKa = 10.38IYY99 pKa = 8.84NTWAGGLDD107 pKa = 4.3FKK109 pKa = 11.63AKK111 pKa = 9.55IAGTGFHH118 pKa = 7.01AGALALVRR126 pKa = 11.84LPPNVDD132 pKa = 3.14PATVSDD138 pKa = 4.13VNKK141 pKa = 7.92FTCFEE146 pKa = 4.22YY147 pKa = 10.73EE148 pKa = 4.06IIDD151 pKa = 4.06PKK153 pKa = 11.13LLEE156 pKa = 4.4CVSRR160 pKa = 11.84SVMDD164 pKa = 3.45QRR166 pKa = 11.84NTMYY170 pKa = 10.36HH171 pKa = 5.62YY172 pKa = 11.06NPYY175 pKa = 10.68DD176 pKa = 3.32ATNYY180 pKa = 10.63NSFGGNFCIFVLQALNTSSSGSTSISVQIFSKK212 pKa = 10.59ASEE215 pKa = 4.44DD216 pKa = 3.97FEE218 pKa = 4.38MLQIIPPNITTLSSTT233 pKa = 3.75
Molecular weight: 24.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLY1|A0A1L3KLY1_9VIRU Uncharacterized protein OS=Hubei picorna-like virus 77 OX=1923161 PE=4 SV=1
MM1 pKa = 7.06FQYY4 pKa = 10.48PHH6 pKa = 7.36LVRR9 pKa = 11.84PNPSNFIKK17 pKa = 10.27IYY19 pKa = 9.87VYY21 pKa = 10.44QIVHH25 pKa = 6.52LRR27 pKa = 11.84LIMYY31 pKa = 7.61LTNRR35 pKa = 11.84YY36 pKa = 8.66AHH38 pKa = 7.23LLPHH42 pKa = 6.63PLGRR46 pKa = 11.84TPRR49 pKa = 11.84DD50 pKa = 3.45PNSVFTSRR58 pKa = 11.84DD59 pKa = 3.32LPLTPLTTNRR69 pKa = 11.84PRR71 pKa = 11.84NISHH75 pKa = 6.62INEE78 pKa = 4.42SVSSPSRR85 pKa = 11.84YY86 pKa = 9.79SSLSNHH92 pKa = 6.41GPLTHH97 pKa = 6.79SSYY100 pKa = 10.6ATASTPSIHH109 pKa = 6.49SVPSRR114 pKa = 11.84SPSSVFSRR122 pKa = 11.84ANSSSSRR129 pKa = 11.84NSSSIFSRR137 pKa = 11.84SGSQHH142 pKa = 4.48STVPHH147 pKa = 6.81RR148 pKa = 11.84LPHH151 pKa = 5.21NQIVTRR157 pKa = 11.84ATHH160 pKa = 5.99PMSTLTHH167 pKa = 6.71PLSLVPFVGSNSKK180 pKa = 10.21PRR182 pKa = 11.84VDD184 pKa = 4.72FPSHH188 pKa = 6.71KK189 pKa = 9.37YY190 pKa = 10.51SKK192 pKa = 11.05VSIQDD197 pKa = 3.32VTPTKK202 pKa = 10.62SKK204 pKa = 11.54SNFLGDD210 pKa = 3.58AMHH213 pKa = 6.97GAGSGFFAGAGLATGQIYY231 pKa = 8.08STNRR235 pKa = 11.84KK236 pKa = 8.94AKK238 pKa = 10.52AIEE241 pKa = 4.04QYY243 pKa = 10.96NADD246 pKa = 3.56KK247 pKa = 10.85VAAYY251 pKa = 7.4EE252 pKa = 4.09KK253 pKa = 10.65SGLPSYY259 pKa = 9.41MAYY262 pKa = 10.91SNTAPSPPVTATIQGNIVHH281 pKa = 6.18KK282 pKa = 9.32TGNSFQSIGNQPSSYY297 pKa = 10.38LSQASGFANMM307 pKa = 4.8
MM1 pKa = 7.06FQYY4 pKa = 10.48PHH6 pKa = 7.36LVRR9 pKa = 11.84PNPSNFIKK17 pKa = 10.27IYY19 pKa = 9.87VYY21 pKa = 10.44QIVHH25 pKa = 6.52LRR27 pKa = 11.84LIMYY31 pKa = 7.61LTNRR35 pKa = 11.84YY36 pKa = 8.66AHH38 pKa = 7.23LLPHH42 pKa = 6.63PLGRR46 pKa = 11.84TPRR49 pKa = 11.84DD50 pKa = 3.45PNSVFTSRR58 pKa = 11.84DD59 pKa = 3.32LPLTPLTTNRR69 pKa = 11.84PRR71 pKa = 11.84NISHH75 pKa = 6.62INEE78 pKa = 4.42SVSSPSRR85 pKa = 11.84YY86 pKa = 9.79SSLSNHH92 pKa = 6.41GPLTHH97 pKa = 6.79SSYY100 pKa = 10.6ATASTPSIHH109 pKa = 6.49SVPSRR114 pKa = 11.84SPSSVFSRR122 pKa = 11.84ANSSSSRR129 pKa = 11.84NSSSIFSRR137 pKa = 11.84SGSQHH142 pKa = 4.48STVPHH147 pKa = 6.81RR148 pKa = 11.84LPHH151 pKa = 5.21NQIVTRR157 pKa = 11.84ATHH160 pKa = 5.99PMSTLTHH167 pKa = 6.71PLSLVPFVGSNSKK180 pKa = 10.21PRR182 pKa = 11.84VDD184 pKa = 4.72FPSHH188 pKa = 6.71KK189 pKa = 9.37YY190 pKa = 10.51SKK192 pKa = 11.05VSIQDD197 pKa = 3.32VTPTKK202 pKa = 10.62SKK204 pKa = 11.54SNFLGDD210 pKa = 3.58AMHH213 pKa = 6.97GAGSGFFAGAGLATGQIYY231 pKa = 8.08STNRR235 pKa = 11.84KK236 pKa = 8.94AKK238 pKa = 10.52AIEE241 pKa = 4.04QYY243 pKa = 10.96NADD246 pKa = 3.56KK247 pKa = 10.85VAAYY251 pKa = 7.4EE252 pKa = 4.09KK253 pKa = 10.65SGLPSYY259 pKa = 9.41MAYY262 pKa = 10.91SNTAPSPPVTATIQGNIVHH281 pKa = 6.18KK282 pKa = 9.32TGNSFQSIGNQPSSYY297 pKa = 10.38LSQASGFANMM307 pKa = 4.8
Molecular weight: 33.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2802 |
233 |
2029 |
700.5 |
78.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.103 ± 1.065 | 1.499 ± 0.419 |
6.103 ± 1.217 | 3.747 ± 0.855 |
4.996 ± 0.343 | 4.889 ± 0.173 |
3.069 ± 0.476 | 5.996 ± 0.319 |
5.318 ± 0.781 | 9.35 ± 0.829 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.856 ± 0.098 | 5.746 ± 0.329 |
6.067 ± 0.979 | 3.319 ± 0.155 |
4.033 ± 0.502 | 8.708 ± 2.744 |
7.423 ± 0.275 | 6.567 ± 0.794 |
0.714 ± 0.253 | 4.497 ± 0.346 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
