
Lepeophtheirus salmonis rhabdovirus 127
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Caligrhavirus; Lepeophtheirus caligrhavirus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
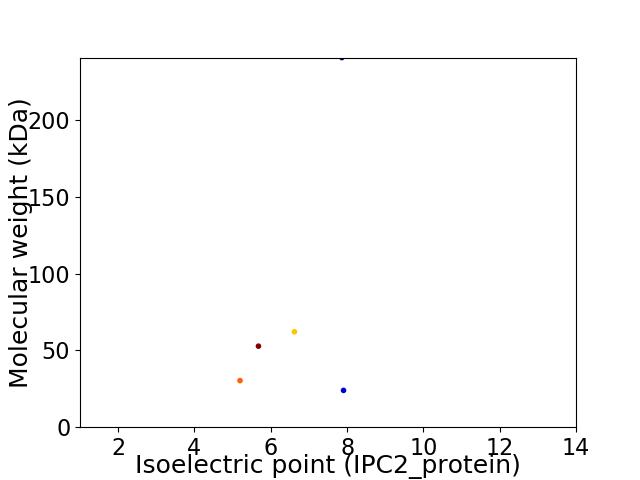
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
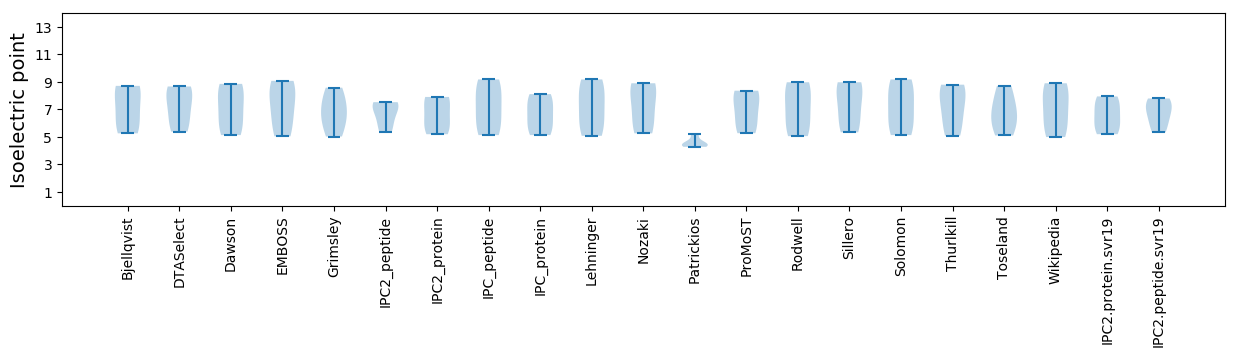
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1EBK8|A0A0A1EBK8_9RHAB GDP polyribonucleotidyltransferase OS=Lepeophtheirus salmonis rhabdovirus 127 OX=1573761 PE=4 SV=1
MM1 pKa = 6.7SQKK4 pKa = 10.08VSNVGAILDD13 pKa = 4.03KK14 pKa = 10.64GVSMVRR20 pKa = 11.84AEE22 pKa = 3.95GRR24 pKa = 11.84RR25 pKa = 11.84AVEE28 pKa = 4.27DD29 pKa = 3.89LGEE32 pKa = 4.15EE33 pKa = 4.42SPDD36 pKa = 3.09EE37 pKa = 5.09DD38 pKa = 4.11YY39 pKa = 11.35QEE41 pKa = 4.69HH42 pKa = 6.49PKK44 pKa = 10.73KK45 pKa = 10.34PFSSTFGTWHH55 pKa = 6.3QKK57 pKa = 8.47FQEE60 pKa = 5.08DD61 pKa = 3.59INDD64 pKa = 3.43WGEE67 pKa = 3.63QRR69 pKa = 11.84EE70 pKa = 4.4IQGLDD75 pKa = 3.14PEE77 pKa = 4.56PRR79 pKa = 11.84STYY82 pKa = 10.94EE83 pKa = 3.66MLRR86 pKa = 11.84DD87 pKa = 3.71PGEE90 pKa = 4.41EE91 pKa = 3.58INIYY95 pKa = 10.23PDD97 pKa = 3.38TDD99 pKa = 3.78LSNMGSQGPEE109 pKa = 3.71VCDD112 pKa = 3.63LQDD115 pKa = 3.18FRR117 pKa = 11.84TVLDD121 pKa = 4.97EE122 pKa = 4.02ISLRR126 pKa = 11.84YY127 pKa = 9.51NIPRR131 pKa = 11.84LIIKK135 pKa = 9.84RR136 pKa = 11.84DD137 pKa = 3.23AGRR140 pKa = 11.84ITWGEE145 pKa = 3.87AGTGEE150 pKa = 4.36EE151 pKa = 4.33PKK153 pKa = 10.27IPQDD157 pKa = 3.27VHH159 pKa = 7.79LVEE162 pKa = 5.22EE163 pKa = 4.81INVKK167 pKa = 10.34PSIGYY172 pKa = 7.41PKK174 pKa = 10.2KK175 pKa = 9.94PKK177 pKa = 10.46ASINEE182 pKa = 4.01TRR184 pKa = 11.84EE185 pKa = 4.05RR186 pKa = 11.84NSLEE190 pKa = 3.42MALIEE195 pKa = 4.68KK196 pKa = 9.8RR197 pKa = 11.84YY198 pKa = 9.33HH199 pKa = 5.51VLGIFDD205 pKa = 3.83RR206 pKa = 11.84TKK208 pKa = 10.65KK209 pKa = 10.42LSLGIPPIQGVNRR222 pKa = 11.84LLADD226 pKa = 3.14IWGEE230 pKa = 4.08EE231 pKa = 3.98KK232 pKa = 11.18AMDD235 pKa = 3.97DD236 pKa = 4.47MIRR239 pKa = 11.84EE240 pKa = 5.01LYY242 pKa = 9.58RR243 pKa = 11.84HH244 pKa = 6.24HH245 pKa = 7.29PNMRR249 pKa = 11.84RR250 pKa = 11.84LSRR253 pKa = 11.84SFNFKK258 pKa = 10.47FIQEE262 pKa = 4.23SS263 pKa = 3.44
MM1 pKa = 6.7SQKK4 pKa = 10.08VSNVGAILDD13 pKa = 4.03KK14 pKa = 10.64GVSMVRR20 pKa = 11.84AEE22 pKa = 3.95GRR24 pKa = 11.84RR25 pKa = 11.84AVEE28 pKa = 4.27DD29 pKa = 3.89LGEE32 pKa = 4.15EE33 pKa = 4.42SPDD36 pKa = 3.09EE37 pKa = 5.09DD38 pKa = 4.11YY39 pKa = 11.35QEE41 pKa = 4.69HH42 pKa = 6.49PKK44 pKa = 10.73KK45 pKa = 10.34PFSSTFGTWHH55 pKa = 6.3QKK57 pKa = 8.47FQEE60 pKa = 5.08DD61 pKa = 3.59INDD64 pKa = 3.43WGEE67 pKa = 3.63QRR69 pKa = 11.84EE70 pKa = 4.4IQGLDD75 pKa = 3.14PEE77 pKa = 4.56PRR79 pKa = 11.84STYY82 pKa = 10.94EE83 pKa = 3.66MLRR86 pKa = 11.84DD87 pKa = 3.71PGEE90 pKa = 4.41EE91 pKa = 3.58INIYY95 pKa = 10.23PDD97 pKa = 3.38TDD99 pKa = 3.78LSNMGSQGPEE109 pKa = 3.71VCDD112 pKa = 3.63LQDD115 pKa = 3.18FRR117 pKa = 11.84TVLDD121 pKa = 4.97EE122 pKa = 4.02ISLRR126 pKa = 11.84YY127 pKa = 9.51NIPRR131 pKa = 11.84LIIKK135 pKa = 9.84RR136 pKa = 11.84DD137 pKa = 3.23AGRR140 pKa = 11.84ITWGEE145 pKa = 3.87AGTGEE150 pKa = 4.36EE151 pKa = 4.33PKK153 pKa = 10.27IPQDD157 pKa = 3.27VHH159 pKa = 7.79LVEE162 pKa = 5.22EE163 pKa = 4.81INVKK167 pKa = 10.34PSIGYY172 pKa = 7.41PKK174 pKa = 10.2KK175 pKa = 9.94PKK177 pKa = 10.46ASINEE182 pKa = 4.01TRR184 pKa = 11.84EE185 pKa = 4.05RR186 pKa = 11.84NSLEE190 pKa = 3.42MALIEE195 pKa = 4.68KK196 pKa = 9.8RR197 pKa = 11.84YY198 pKa = 9.33HH199 pKa = 5.51VLGIFDD205 pKa = 3.83RR206 pKa = 11.84TKK208 pKa = 10.65KK209 pKa = 10.42LSLGIPPIQGVNRR222 pKa = 11.84LLADD226 pKa = 3.14IWGEE230 pKa = 4.08EE231 pKa = 3.98KK232 pKa = 11.18AMDD235 pKa = 3.97DD236 pKa = 4.47MIRR239 pKa = 11.84EE240 pKa = 5.01LYY242 pKa = 9.58RR243 pKa = 11.84HH244 pKa = 6.24HH245 pKa = 7.29PNMRR249 pKa = 11.84RR250 pKa = 11.84LSRR253 pKa = 11.84SFNFKK258 pKa = 10.47FIQEE262 pKa = 4.23SS263 pKa = 3.44
Molecular weight: 30.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1E924|A0A0A1E924_9RHAB Phosphoprotein OS=Lepeophtheirus salmonis rhabdovirus 127 OX=1573761 PE=4 SV=1
MM1 pKa = 7.23LRR3 pKa = 11.84KK4 pKa = 9.28AALALRR10 pKa = 11.84GGNKK14 pKa = 10.01EE15 pKa = 3.92EE16 pKa = 4.63DD17 pKa = 3.4RR18 pKa = 11.84HH19 pKa = 6.28PVTSWSSPIQSPSAPMDD36 pKa = 3.57SRR38 pKa = 11.84LLTVNLKK45 pKa = 8.37VTAHH49 pKa = 5.46MSVRR53 pKa = 11.84TTNKK57 pKa = 9.82NIVFSDD63 pKa = 3.58VLLACTDD70 pKa = 3.09IMDD73 pKa = 4.36TYY75 pKa = 10.45DD76 pKa = 4.46GPMEE80 pKa = 4.01TLGFYY85 pKa = 10.14WIMILVLVKK94 pKa = 10.36RR95 pKa = 11.84LKK97 pKa = 10.44SLRR100 pKa = 11.84GNEE103 pKa = 3.9GTQDD107 pKa = 3.58FVSSVSEE114 pKa = 4.31PISFLTDD121 pKa = 3.13ISSPYY126 pKa = 10.71LKK128 pKa = 9.76TFPTTAFSFGRR139 pKa = 11.84EE140 pKa = 3.69FRR142 pKa = 11.84AGKK145 pKa = 10.24VSGNASVSFSEE156 pKa = 4.35TRR158 pKa = 11.84AVTTSLEE165 pKa = 4.81LILQSLGLVNDD176 pKa = 4.32HH177 pKa = 6.86LGSALSALKK186 pKa = 10.43IPAEE190 pKa = 4.3RR191 pKa = 11.84IKK193 pKa = 10.98GSLVIHH199 pKa = 6.13MALRR203 pKa = 11.84DD204 pKa = 4.16SFSHH208 pKa = 6.19IQPATLKK215 pKa = 8.32EE216 pKa = 4.23TSDD219 pKa = 3.41
MM1 pKa = 7.23LRR3 pKa = 11.84KK4 pKa = 9.28AALALRR10 pKa = 11.84GGNKK14 pKa = 10.01EE15 pKa = 3.92EE16 pKa = 4.63DD17 pKa = 3.4RR18 pKa = 11.84HH19 pKa = 6.28PVTSWSSPIQSPSAPMDD36 pKa = 3.57SRR38 pKa = 11.84LLTVNLKK45 pKa = 8.37VTAHH49 pKa = 5.46MSVRR53 pKa = 11.84TTNKK57 pKa = 9.82NIVFSDD63 pKa = 3.58VLLACTDD70 pKa = 3.09IMDD73 pKa = 4.36TYY75 pKa = 10.45DD76 pKa = 4.46GPMEE80 pKa = 4.01TLGFYY85 pKa = 10.14WIMILVLVKK94 pKa = 10.36RR95 pKa = 11.84LKK97 pKa = 10.44SLRR100 pKa = 11.84GNEE103 pKa = 3.9GTQDD107 pKa = 3.58FVSSVSEE114 pKa = 4.31PISFLTDD121 pKa = 3.13ISSPYY126 pKa = 10.71LKK128 pKa = 9.76TFPTTAFSFGRR139 pKa = 11.84EE140 pKa = 3.69FRR142 pKa = 11.84AGKK145 pKa = 10.24VSGNASVSFSEE156 pKa = 4.35TRR158 pKa = 11.84AVTTSLEE165 pKa = 4.81LILQSLGLVNDD176 pKa = 4.32HH177 pKa = 6.86LGSALSALKK186 pKa = 10.43IPAEE190 pKa = 4.3RR191 pKa = 11.84IKK193 pKa = 10.98GSLVIHH199 pKa = 6.13MALRR203 pKa = 11.84DD204 pKa = 4.16SFSHH208 pKa = 6.19IQPATLKK215 pKa = 8.32EE216 pKa = 4.23TSDD219 pKa = 3.41
Molecular weight: 23.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3586 |
219 |
2096 |
717.2 |
82.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.183 ± 0.533 | 1.422 ± 0.382 |
6.191 ± 0.119 | 6.219 ± 0.38 |
4.406 ± 0.151 | 5.8 ± 0.199 |
2.566 ± 0.254 | 6.804 ± 0.199 |
6.219 ± 0.566 | 10.318 ± 0.674 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.566 ± 0.12 | 4.044 ± 0.185 |
4.713 ± 0.508 | 2.817 ± 0.146 |
6.414 ± 0.397 | 8.645 ± 0.411 |
6.079 ± 0.452 | 5.577 ± 0.407 |
2.091 ± 0.185 | 2.928 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
