
Rotavirus I
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
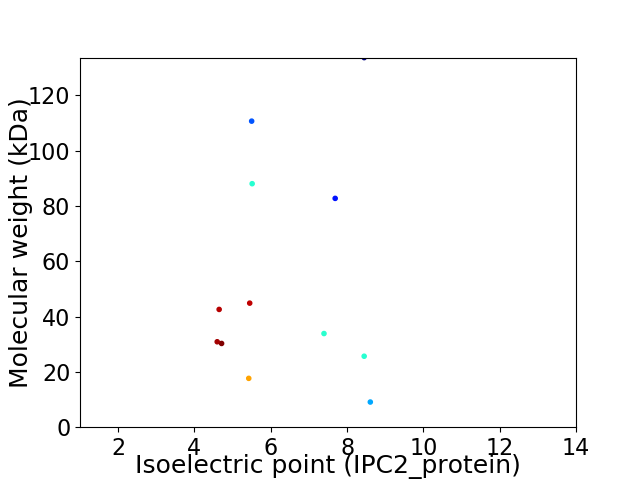
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3JKM5|A0A0E3JKM5_9REOV NSP2 OS=Rotavirus I OX=1637496 PE=4 SV=1
MM1 pKa = 7.71WFLLLLPIVSSTVVIQEE18 pKa = 3.96YY19 pKa = 10.48KK20 pKa = 10.45PVDD23 pKa = 3.41VCIFVYY29 pKa = 10.58ASSGDD34 pKa = 3.6NSSAYY39 pKa = 10.13YY40 pKa = 10.66AFHH43 pKa = 6.3TWMQRR48 pKa = 11.84LQPVLATTYY57 pKa = 9.89HH58 pKa = 5.15VCGVQAVDD66 pKa = 3.45SVDD69 pKa = 3.67VIKK72 pKa = 10.79AVAEE76 pKa = 4.43CSCLNDD82 pKa = 3.74KK83 pKa = 9.63LTDD86 pKa = 3.62VNIVYY91 pKa = 10.37SDD93 pKa = 3.93EE94 pKa = 4.43PQSTLATFIGNQNTCTKK111 pKa = 10.2LQVRR115 pKa = 11.84KK116 pKa = 9.29AAYY119 pKa = 8.15IHH121 pKa = 6.39LNKK124 pKa = 10.2SEE126 pKa = 3.93EE127 pKa = 3.75FFIYY131 pKa = 9.99GKK133 pKa = 10.24RR134 pKa = 11.84DD135 pKa = 3.2LKK137 pKa = 11.26LCFLSSSYY145 pKa = 11.12LGLDD149 pKa = 3.5CDD151 pKa = 4.59SANPTTWPTIDD162 pKa = 4.41YY163 pKa = 9.03NIQSIIHH170 pKa = 6.5EE171 pKa = 4.44KK172 pKa = 10.05PIIYY176 pKa = 9.2DD177 pKa = 3.32IPEE180 pKa = 3.38ITYY183 pKa = 10.66AVGIYY188 pKa = 10.15PEE190 pKa = 4.91ADD192 pKa = 2.77PTYY195 pKa = 8.93ICKK198 pKa = 10.27RR199 pKa = 11.84VDD201 pKa = 3.51DD202 pKa = 4.45NGINPTYY209 pKa = 10.5FFAGDD214 pKa = 4.23NSIQWGDD221 pKa = 3.35QMARR225 pKa = 11.84EE226 pKa = 4.38INWGNVWNKK235 pKa = 9.36VVSVSQALYY244 pKa = 10.42KK245 pKa = 10.5ILDD248 pKa = 3.52LFFNEE253 pKa = 4.69KK254 pKa = 9.99EE255 pKa = 4.28RR256 pKa = 11.84IEE258 pKa = 3.99QQPRR262 pKa = 11.84YY263 pKa = 9.27EE264 pKa = 3.96LL265 pKa = 3.9
MM1 pKa = 7.71WFLLLLPIVSSTVVIQEE18 pKa = 3.96YY19 pKa = 10.48KK20 pKa = 10.45PVDD23 pKa = 3.41VCIFVYY29 pKa = 10.58ASSGDD34 pKa = 3.6NSSAYY39 pKa = 10.13YY40 pKa = 10.66AFHH43 pKa = 6.3TWMQRR48 pKa = 11.84LQPVLATTYY57 pKa = 9.89HH58 pKa = 5.15VCGVQAVDD66 pKa = 3.45SVDD69 pKa = 3.67VIKK72 pKa = 10.79AVAEE76 pKa = 4.43CSCLNDD82 pKa = 3.74KK83 pKa = 9.63LTDD86 pKa = 3.62VNIVYY91 pKa = 10.37SDD93 pKa = 3.93EE94 pKa = 4.43PQSTLATFIGNQNTCTKK111 pKa = 10.2LQVRR115 pKa = 11.84KK116 pKa = 9.29AAYY119 pKa = 8.15IHH121 pKa = 6.39LNKK124 pKa = 10.2SEE126 pKa = 3.93EE127 pKa = 3.75FFIYY131 pKa = 9.99GKK133 pKa = 10.24RR134 pKa = 11.84DD135 pKa = 3.2LKK137 pKa = 11.26LCFLSSSYY145 pKa = 11.12LGLDD149 pKa = 3.5CDD151 pKa = 4.59SANPTTWPTIDD162 pKa = 4.41YY163 pKa = 9.03NIQSIIHH170 pKa = 6.5EE171 pKa = 4.44KK172 pKa = 10.05PIIYY176 pKa = 9.2DD177 pKa = 3.32IPEE180 pKa = 3.38ITYY183 pKa = 10.66AVGIYY188 pKa = 10.15PEE190 pKa = 4.91ADD192 pKa = 2.77PTYY195 pKa = 8.93ICKK198 pKa = 10.27RR199 pKa = 11.84VDD201 pKa = 3.51DD202 pKa = 4.45NGINPTYY209 pKa = 10.5FFAGDD214 pKa = 4.23NSIQWGDD221 pKa = 3.35QMARR225 pKa = 11.84EE226 pKa = 4.38INWGNVWNKK235 pKa = 9.36VVSVSQALYY244 pKa = 10.42KK245 pKa = 10.5ILDD248 pKa = 3.52LFFNEE253 pKa = 4.69KK254 pKa = 9.99EE255 pKa = 4.28RR256 pKa = 11.84IEE258 pKa = 3.99QQPRR262 pKa = 11.84YY263 pKa = 9.27EE264 pKa = 3.96LL265 pKa = 3.9
Molecular weight: 30.27 kDa
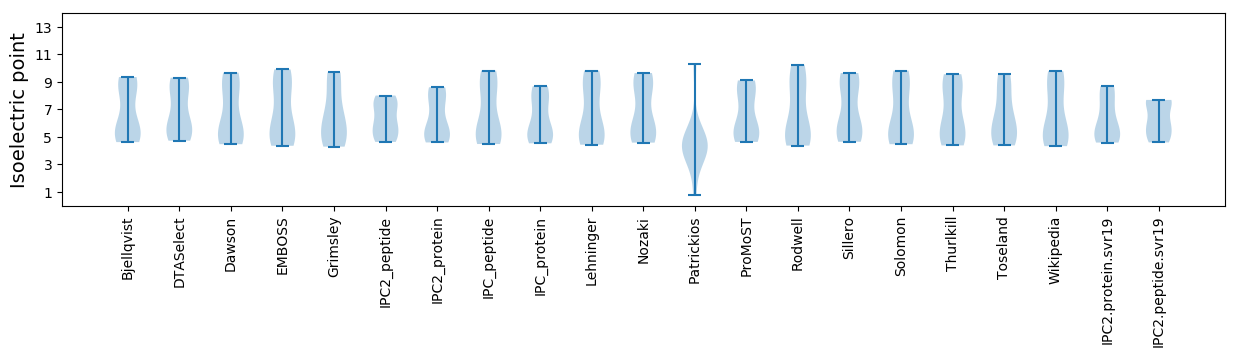
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3M4A3|A0A0E3M4A3_9REOV NSP4 OS=Rotavirus I OX=1637496 PE=4 SV=1
MM1 pKa = 7.33GNTYY5 pKa = 10.4NVHH8 pKa = 6.43NNQQVSSNTVHH19 pKa = 7.19GSGQIHH25 pKa = 6.61SEE27 pKa = 4.05DD28 pKa = 3.58QKK30 pKa = 10.02TSSQITTIVQFSNLSLLFLIALFLFISLLFKK61 pKa = 10.6CDD63 pKa = 3.54KK64 pKa = 10.37NRR66 pKa = 11.84KK67 pKa = 6.34KK68 pKa = 10.72QKK70 pKa = 8.55WNTRR74 pKa = 11.84EE75 pKa = 4.01IIEE78 pKa = 4.19LL79 pKa = 3.69
MM1 pKa = 7.33GNTYY5 pKa = 10.4NVHH8 pKa = 6.43NNQQVSSNTVHH19 pKa = 7.19GSGQIHH25 pKa = 6.61SEE27 pKa = 4.05DD28 pKa = 3.58QKK30 pKa = 10.02TSSQITTIVQFSNLSLLFLIALFLFISLLFKK61 pKa = 10.6CDD63 pKa = 3.54KK64 pKa = 10.37NRR66 pKa = 11.84KK67 pKa = 6.34KK68 pKa = 10.72QKK70 pKa = 8.55WNTRR74 pKa = 11.84EE75 pKa = 4.01IIEE78 pKa = 4.19LL79 pKa = 3.69
Molecular weight: 9.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5698 |
79 |
1162 |
474.8 |
54.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.441 ± 0.676 | 1.211 ± 0.274 |
6.546 ± 0.368 | 5.704 ± 0.511 |
4.581 ± 0.349 | 4.23 ± 0.317 |
1.983 ± 0.237 | 8.424 ± 0.45 |
6.704 ± 0.491 | 8.529 ± 0.333 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.861 ± 0.289 | 6.897 ± 0.491 |
3.703 ± 0.302 | 3.896 ± 0.315 |
4.756 ± 0.316 | 7.687 ± 0.372 |
6.722 ± 0.348 | 5.476 ± 0.312 |
0.86 ± 0.163 | 3.791 ± 0.452 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
