
Lake Sarah-associated circular virus-18
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.94
Get precalculated fractions of proteins
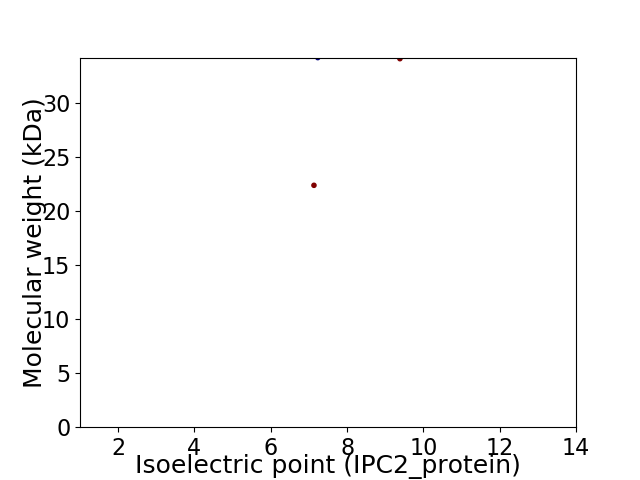
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA83|A0A126GA83_9VIRU Coat protein OS=Lake Sarah-associated circular virus-18 OX=1685744 PE=4 SV=1
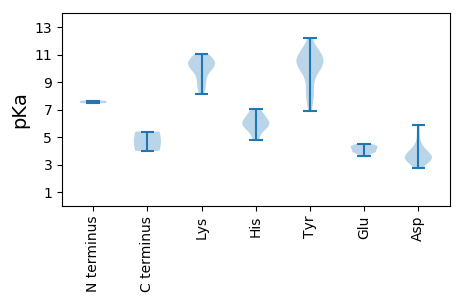
MM1 pKa = 7.46TSLLPTDD8 pKa = 3.91SKK10 pKa = 8.98STRR13 pKa = 11.84WAFTAFEE20 pKa = 4.08SQYY23 pKa = 10.77TLFNEE28 pKa = 4.28MPAFIAQWGWQQEE41 pKa = 4.27VSPTTGKK48 pKa = 9.22PHH50 pKa = 5.4FQGYY54 pKa = 8.55FRR56 pKa = 11.84TTTQVRR62 pKa = 11.84FAYY65 pKa = 10.32LKK67 pKa = 8.74RR68 pKa = 11.84QFPGVHH74 pKa = 6.54LEE76 pKa = 3.9IAKK79 pKa = 10.26DD80 pKa = 3.32WEE82 pKa = 4.41SLKK85 pKa = 10.71KK86 pKa = 10.32YY87 pKa = 10.0CNKK90 pKa = 10.4AEE92 pKa = 4.05TAIPGTQVQATSNFMNMYY110 pKa = 10.46DD111 pKa = 3.93YY112 pKa = 11.19LPWILLQTIDD122 pKa = 3.31TYY124 pKa = 11.45GWQEE128 pKa = 3.63LWDD131 pKa = 4.32DD132 pKa = 3.6SRR134 pKa = 11.84EE135 pKa = 3.97EE136 pKa = 4.12YY137 pKa = 10.06IEE139 pKa = 3.88KK140 pKa = 10.53VMYY143 pKa = 7.85TARR146 pKa = 11.84AHH148 pKa = 5.91IASGNTWASWIIQNPNFKK166 pKa = 10.69QNLKK170 pKa = 10.0EE171 pKa = 3.91NGRR174 pKa = 11.84AILLGFKK181 pKa = 10.02HH182 pKa = 6.08RR183 pKa = 11.84QTDD186 pKa = 3.49RR187 pKa = 11.84QTEE190 pKa = 3.99
MM1 pKa = 7.46TSLLPTDD8 pKa = 3.91SKK10 pKa = 8.98STRR13 pKa = 11.84WAFTAFEE20 pKa = 4.08SQYY23 pKa = 10.77TLFNEE28 pKa = 4.28MPAFIAQWGWQQEE41 pKa = 4.27VSPTTGKK48 pKa = 9.22PHH50 pKa = 5.4FQGYY54 pKa = 8.55FRR56 pKa = 11.84TTTQVRR62 pKa = 11.84FAYY65 pKa = 10.32LKK67 pKa = 8.74RR68 pKa = 11.84QFPGVHH74 pKa = 6.54LEE76 pKa = 3.9IAKK79 pKa = 10.26DD80 pKa = 3.32WEE82 pKa = 4.41SLKK85 pKa = 10.71KK86 pKa = 10.32YY87 pKa = 10.0CNKK90 pKa = 10.4AEE92 pKa = 4.05TAIPGTQVQATSNFMNMYY110 pKa = 10.46DD111 pKa = 3.93YY112 pKa = 11.19LPWILLQTIDD122 pKa = 3.31TYY124 pKa = 11.45GWQEE128 pKa = 3.63LWDD131 pKa = 4.32DD132 pKa = 3.6SRR134 pKa = 11.84EE135 pKa = 3.97EE136 pKa = 4.12YY137 pKa = 10.06IEE139 pKa = 3.88KK140 pKa = 10.53VMYY143 pKa = 7.85TARR146 pKa = 11.84AHH148 pKa = 5.91IASGNTWASWIIQNPNFKK166 pKa = 10.69QNLKK170 pKa = 10.0EE171 pKa = 3.91NGRR174 pKa = 11.84AILLGFKK181 pKa = 10.02HH182 pKa = 6.08RR183 pKa = 11.84QTDD186 pKa = 3.49RR187 pKa = 11.84QTEE190 pKa = 3.99
Molecular weight: 22.38 kDa
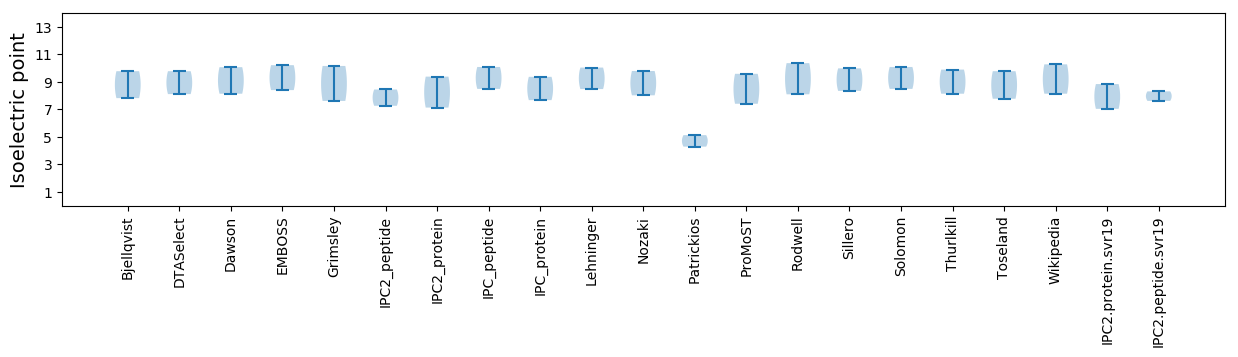
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA83|A0A126GA83_9VIRU Coat protein OS=Lake Sarah-associated circular virus-18 OX=1685744 PE=4 SV=1
MM1 pKa = 7.65PRR3 pKa = 11.84RR4 pKa = 11.84SNRR7 pKa = 11.84LASKK11 pKa = 10.23PRR13 pKa = 11.84LDD15 pKa = 3.41YY16 pKa = 10.95KK17 pKa = 10.71KK18 pKa = 10.29YY19 pKa = 7.86HH20 pKa = 4.79TTGIIVEE27 pKa = 4.25KK28 pKa = 10.55GKK30 pKa = 8.37TRR32 pKa = 11.84AKK34 pKa = 9.88RR35 pKa = 11.84VQSARR40 pKa = 11.84ATGALATAIKK50 pKa = 10.34KK51 pKa = 8.11VVRR54 pKa = 11.84GQAEE58 pKa = 4.28TKK60 pKa = 9.06MATWYY65 pKa = 10.86SGGTNPLGVGARR77 pKa = 11.84SNWAAEE83 pKa = 3.85PHH85 pKa = 5.85NAQITSNTTDD95 pKa = 2.71IMRR98 pKa = 11.84LIPVVAVGTGDD109 pKa = 3.32NQRR112 pKa = 11.84IGEE115 pKa = 4.5RR116 pKa = 11.84ISPSSFIVNGVVSINLSNVLVNVAPLDD143 pKa = 3.69IVAVIYY149 pKa = 9.7VLQHH153 pKa = 6.57KK154 pKa = 8.49SLKK157 pKa = 9.63TYY159 pKa = 11.07NSLQTVTQPGPPIVQSGNDD178 pKa = 3.63FSQLLLTGEE187 pKa = 4.43GDD189 pKa = 3.69TVGFDD194 pKa = 3.04GHH196 pKa = 7.03PYY198 pKa = 10.09DD199 pKa = 5.86ASLQVADD206 pKa = 5.06QYY208 pKa = 12.18YY209 pKa = 10.36KK210 pKa = 11.06LCAKK214 pKa = 10.32KK215 pKa = 10.54LIPLRR220 pKa = 11.84YY221 pKa = 9.0AGSNVVPTGSPIPGGAGVVSVANSHH246 pKa = 6.05TYY248 pKa = 8.15SARR251 pKa = 11.84YY252 pKa = 6.87TFNLKK257 pKa = 9.91KK258 pKa = 10.12HH259 pKa = 5.83IPSVLKK265 pKa = 9.58YY266 pKa = 10.17QEE268 pKa = 4.35SAVTTGQPGDD278 pKa = 4.21PLNSSIFMCVGYY290 pKa = 10.57YY291 pKa = 10.48RR292 pKa = 11.84LDD294 pKa = 3.38QTVPGAAAFLSNSYY308 pKa = 10.6VATLKK313 pKa = 10.99YY314 pKa = 10.8KK315 pKa = 10.96DD316 pKa = 3.41MM317 pKa = 5.4
MM1 pKa = 7.65PRR3 pKa = 11.84RR4 pKa = 11.84SNRR7 pKa = 11.84LASKK11 pKa = 10.23PRR13 pKa = 11.84LDD15 pKa = 3.41YY16 pKa = 10.95KK17 pKa = 10.71KK18 pKa = 10.29YY19 pKa = 7.86HH20 pKa = 4.79TTGIIVEE27 pKa = 4.25KK28 pKa = 10.55GKK30 pKa = 8.37TRR32 pKa = 11.84AKK34 pKa = 9.88RR35 pKa = 11.84VQSARR40 pKa = 11.84ATGALATAIKK50 pKa = 10.34KK51 pKa = 8.11VVRR54 pKa = 11.84GQAEE58 pKa = 4.28TKK60 pKa = 9.06MATWYY65 pKa = 10.86SGGTNPLGVGARR77 pKa = 11.84SNWAAEE83 pKa = 3.85PHH85 pKa = 5.85NAQITSNTTDD95 pKa = 2.71IMRR98 pKa = 11.84LIPVVAVGTGDD109 pKa = 3.32NQRR112 pKa = 11.84IGEE115 pKa = 4.5RR116 pKa = 11.84ISPSSFIVNGVVSINLSNVLVNVAPLDD143 pKa = 3.69IVAVIYY149 pKa = 9.7VLQHH153 pKa = 6.57KK154 pKa = 8.49SLKK157 pKa = 9.63TYY159 pKa = 11.07NSLQTVTQPGPPIVQSGNDD178 pKa = 3.63FSQLLLTGEE187 pKa = 4.43GDD189 pKa = 3.69TVGFDD194 pKa = 3.04GHH196 pKa = 7.03PYY198 pKa = 10.09DD199 pKa = 5.86ASLQVADD206 pKa = 5.06QYY208 pKa = 12.18YY209 pKa = 10.36KK210 pKa = 11.06LCAKK214 pKa = 10.32KK215 pKa = 10.54LIPLRR220 pKa = 11.84YY221 pKa = 9.0AGSNVVPTGSPIPGGAGVVSVANSHH246 pKa = 6.05TYY248 pKa = 8.15SARR251 pKa = 11.84YY252 pKa = 6.87TFNLKK257 pKa = 9.91KK258 pKa = 10.12HH259 pKa = 5.83IPSVLKK265 pKa = 9.58YY266 pKa = 10.17QEE268 pKa = 4.35SAVTTGQPGDD278 pKa = 4.21PLNSSIFMCVGYY290 pKa = 10.57YY291 pKa = 10.48RR292 pKa = 11.84LDD294 pKa = 3.38QTVPGAAAFLSNSYY308 pKa = 10.6VATLKK313 pKa = 10.99YY314 pKa = 10.8KK315 pKa = 10.96DD316 pKa = 3.41MM317 pKa = 5.4
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
507 |
190 |
317 |
253.5 |
28.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.284 ± 0.552 | 0.592 ± 0.039 |
3.748 ± 0.038 | 3.55 ± 1.666 |
3.353 ± 1.468 | 7.101 ± 1.424 |
1.972 ± 0.08 | 5.325 ± 0.038 |
5.917 ± 0.077 | 7.298 ± 0.275 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.397 | 5.128 ± 0.236 |
5.325 ± 0.672 | 5.72 ± 1.31 |
4.734 ± 0.002 | 7.298 ± 1.226 |
8.481 ± 0.915 | 7.101 ± 2.693 |
2.17 ± 1.547 | 4.931 ± 0.117 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
