
Zygosaccharomyces bailii virus Z
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; Zybavirus
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
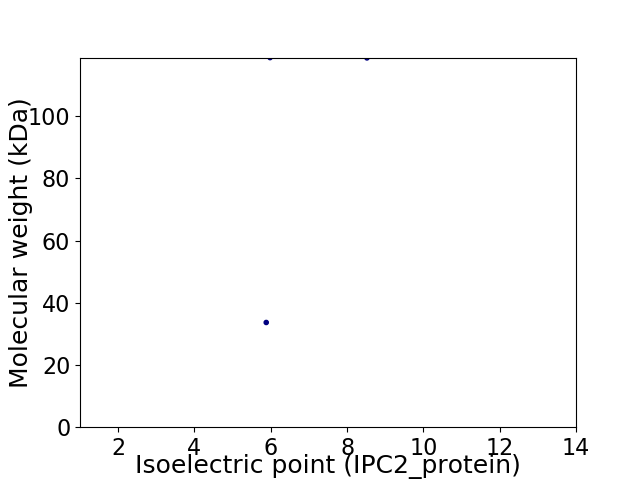
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9J0H5|Q9J0H5_9VIRU Major capsid protein OS=Zygosaccharomyces bailii virus Z OX=114871 GN=CAP PE=4 SV=1
MM1 pKa = 7.52EE2 pKa = 6.26KK3 pKa = 10.3DD4 pKa = 3.74GKK6 pKa = 10.83SSTWSSFSTALSEE19 pKa = 4.18IQKK22 pKa = 9.92EE23 pKa = 4.16RR24 pKa = 11.84VPGLEE29 pKa = 4.2EE30 pKa = 3.61KK31 pKa = 10.3TSIDD35 pKa = 3.65FEE37 pKa = 4.86VDD39 pKa = 2.78GKK41 pKa = 10.88RR42 pKa = 11.84YY43 pKa = 10.44SLVSSRR49 pKa = 11.84KK50 pKa = 7.12PTDD53 pKa = 3.19RR54 pKa = 11.84EE55 pKa = 4.14VEE57 pKa = 4.27LKK59 pKa = 10.85SLIPSAILTEE69 pKa = 3.86QLLSNEE75 pKa = 4.62HH76 pKa = 5.66IGKK79 pKa = 9.16YY80 pKa = 7.77FEE82 pKa = 5.29RR83 pKa = 11.84IRR85 pKa = 11.84KK86 pKa = 8.68ALIVKK91 pKa = 7.61NTQADD96 pKa = 4.03LVEE99 pKa = 4.58LLNSFTTVLTNIHH112 pKa = 5.73QVSLYY117 pKa = 10.07IEE119 pKa = 4.22HH120 pKa = 7.08HH121 pKa = 7.16PDD123 pKa = 2.76TTAVAKK129 pKa = 10.8SFEE132 pKa = 4.15NYY134 pKa = 9.55KK135 pKa = 10.5LKK137 pKa = 10.72KK138 pKa = 10.23DD139 pKa = 3.6LSKK142 pKa = 10.84WKK144 pKa = 10.6LDD146 pKa = 3.5MLNSLSTVDD155 pKa = 3.69GRR157 pKa = 11.84VQIHH161 pKa = 5.45VNEE164 pKa = 4.04ILAMFHH170 pKa = 5.83GVKK173 pKa = 10.35NAAQKK178 pKa = 9.8DD179 pKa = 3.73AQMKK183 pKa = 8.82EE184 pKa = 3.74EE185 pKa = 4.17KK186 pKa = 10.08ARR188 pKa = 11.84KK189 pKa = 9.1LMKK192 pKa = 10.26QQVEE196 pKa = 4.02LARR199 pKa = 11.84QEE201 pKa = 3.82FEE203 pKa = 3.98EE204 pKa = 4.77RR205 pKa = 11.84IKK207 pKa = 10.65HH208 pKa = 4.57VRR210 pKa = 11.84PFQGLMLLTGSFKK223 pKa = 10.06EE224 pKa = 4.09QSDD227 pKa = 3.98RR228 pKa = 11.84VHH230 pKa = 6.51EE231 pKa = 4.2MARR234 pKa = 11.84QEE236 pKa = 4.26WMSLPDD242 pKa = 3.62TEE244 pKa = 5.47KK245 pKa = 11.18SSDD248 pKa = 3.25YY249 pKa = 11.02DD250 pKa = 2.99AWAQAWWATRR260 pKa = 11.84KK261 pKa = 10.32DD262 pKa = 3.67GEE264 pKa = 4.23IFAVLEE270 pKa = 4.27STVMTEE276 pKa = 3.66EE277 pKa = 3.76FLEE280 pKa = 4.71FFSQHH285 pKa = 4.2WLFRR289 pKa = 11.84GG290 pKa = 3.45
MM1 pKa = 7.52EE2 pKa = 6.26KK3 pKa = 10.3DD4 pKa = 3.74GKK6 pKa = 10.83SSTWSSFSTALSEE19 pKa = 4.18IQKK22 pKa = 9.92EE23 pKa = 4.16RR24 pKa = 11.84VPGLEE29 pKa = 4.2EE30 pKa = 3.61KK31 pKa = 10.3TSIDD35 pKa = 3.65FEE37 pKa = 4.86VDD39 pKa = 2.78GKK41 pKa = 10.88RR42 pKa = 11.84YY43 pKa = 10.44SLVSSRR49 pKa = 11.84KK50 pKa = 7.12PTDD53 pKa = 3.19RR54 pKa = 11.84EE55 pKa = 4.14VEE57 pKa = 4.27LKK59 pKa = 10.85SLIPSAILTEE69 pKa = 3.86QLLSNEE75 pKa = 4.62HH76 pKa = 5.66IGKK79 pKa = 9.16YY80 pKa = 7.77FEE82 pKa = 5.29RR83 pKa = 11.84IRR85 pKa = 11.84KK86 pKa = 8.68ALIVKK91 pKa = 7.61NTQADD96 pKa = 4.03LVEE99 pKa = 4.58LLNSFTTVLTNIHH112 pKa = 5.73QVSLYY117 pKa = 10.07IEE119 pKa = 4.22HH120 pKa = 7.08HH121 pKa = 7.16PDD123 pKa = 2.76TTAVAKK129 pKa = 10.8SFEE132 pKa = 4.15NYY134 pKa = 9.55KK135 pKa = 10.5LKK137 pKa = 10.72KK138 pKa = 10.23DD139 pKa = 3.6LSKK142 pKa = 10.84WKK144 pKa = 10.6LDD146 pKa = 3.5MLNSLSTVDD155 pKa = 3.69GRR157 pKa = 11.84VQIHH161 pKa = 5.45VNEE164 pKa = 4.04ILAMFHH170 pKa = 5.83GVKK173 pKa = 10.35NAAQKK178 pKa = 9.8DD179 pKa = 3.73AQMKK183 pKa = 8.82EE184 pKa = 3.74EE185 pKa = 4.17KK186 pKa = 10.08ARR188 pKa = 11.84KK189 pKa = 9.1LMKK192 pKa = 10.26QQVEE196 pKa = 4.02LARR199 pKa = 11.84QEE201 pKa = 3.82FEE203 pKa = 3.98EE204 pKa = 4.77RR205 pKa = 11.84IKK207 pKa = 10.65HH208 pKa = 4.57VRR210 pKa = 11.84PFQGLMLLTGSFKK223 pKa = 10.06EE224 pKa = 4.09QSDD227 pKa = 3.98RR228 pKa = 11.84VHH230 pKa = 6.51EE231 pKa = 4.2MARR234 pKa = 11.84QEE236 pKa = 4.26WMSLPDD242 pKa = 3.62TEE244 pKa = 5.47KK245 pKa = 11.18SSDD248 pKa = 3.25YY249 pKa = 11.02DD250 pKa = 2.99AWAQAWWATRR260 pKa = 11.84KK261 pKa = 10.32DD262 pKa = 3.67GEE264 pKa = 4.23IFAVLEE270 pKa = 4.27STVMTEE276 pKa = 3.66EE277 pKa = 3.76FLEE280 pKa = 4.71FFSQHH285 pKa = 4.2WLFRR289 pKa = 11.84GG290 pKa = 3.45
Molecular weight: 33.68 kDa
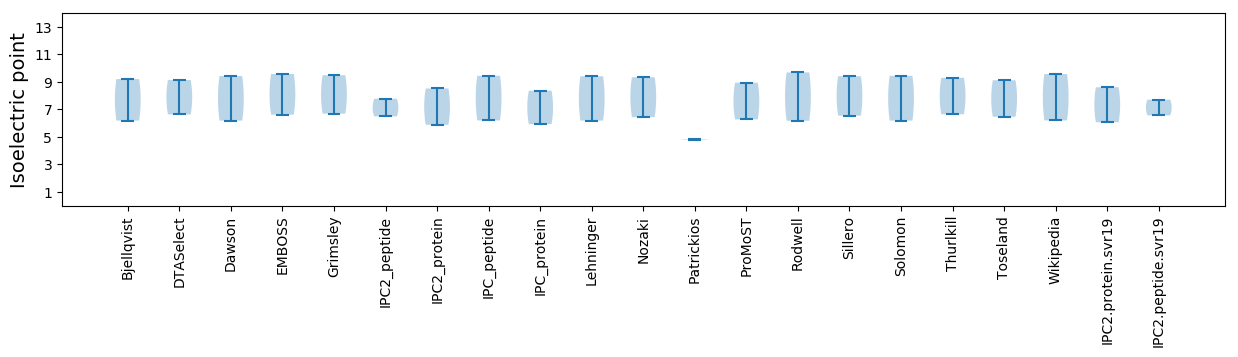
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9J0H5|Q9J0H5_9VIRU Major capsid protein OS=Zygosaccharomyces bailii virus Z OX=114871 GN=CAP PE=4 SV=1
MM1 pKa = 7.52EE2 pKa = 6.26KK3 pKa = 10.3DD4 pKa = 3.74GKK6 pKa = 10.83SSTWSSFSTALSEE19 pKa = 4.18IQKK22 pKa = 9.92EE23 pKa = 4.16RR24 pKa = 11.84VPGLEE29 pKa = 4.2EE30 pKa = 3.61KK31 pKa = 10.3TSIDD35 pKa = 3.65FEE37 pKa = 4.86VDD39 pKa = 2.78GKK41 pKa = 10.88RR42 pKa = 11.84YY43 pKa = 10.44SLVSSRR49 pKa = 11.84KK50 pKa = 7.12PTDD53 pKa = 3.19RR54 pKa = 11.84EE55 pKa = 4.14VEE57 pKa = 4.27LKK59 pKa = 10.85SLIPSAILTEE69 pKa = 3.86QLLSNEE75 pKa = 4.62HH76 pKa = 5.66IGKK79 pKa = 9.16YY80 pKa = 7.77FEE82 pKa = 5.29RR83 pKa = 11.84IRR85 pKa = 11.84KK86 pKa = 8.68ALIVKK91 pKa = 7.61NTQADD96 pKa = 4.03LVEE99 pKa = 4.58LLNSFTTVLTNIHH112 pKa = 5.73QVSLYY117 pKa = 10.07IEE119 pKa = 4.22HH120 pKa = 7.08HH121 pKa = 7.16PDD123 pKa = 2.76TTAVAKK129 pKa = 10.8SFEE132 pKa = 4.15NYY134 pKa = 9.55KK135 pKa = 10.5LKK137 pKa = 10.72KK138 pKa = 10.23DD139 pKa = 3.6LSKK142 pKa = 10.84WKK144 pKa = 10.6LDD146 pKa = 3.5MLNSLSTVDD155 pKa = 3.69GRR157 pKa = 11.84VQIHH161 pKa = 5.45VNEE164 pKa = 4.04ILAMFHH170 pKa = 5.83GVKK173 pKa = 10.35NAAQKK178 pKa = 9.8DD179 pKa = 3.73AQMKK183 pKa = 8.82EE184 pKa = 3.74EE185 pKa = 4.17KK186 pKa = 10.08ARR188 pKa = 11.84KK189 pKa = 9.1LMKK192 pKa = 10.26QQVEE196 pKa = 4.02LARR199 pKa = 11.84QEE201 pKa = 3.82FEE203 pKa = 3.98EE204 pKa = 4.77RR205 pKa = 11.84IKK207 pKa = 10.65HH208 pKa = 4.57VRR210 pKa = 11.84PFQGLMLLTGSFKK223 pKa = 10.06EE224 pKa = 4.09QSDD227 pKa = 3.98RR228 pKa = 11.84VHH230 pKa = 6.51EE231 pKa = 4.2MARR234 pKa = 11.84QEE236 pKa = 4.26WMSLPDD242 pKa = 3.62TEE244 pKa = 5.47KK245 pKa = 11.18SSDD248 pKa = 3.25YY249 pKa = 11.02DD250 pKa = 2.99AWAQAWWATRR260 pKa = 11.84KK261 pKa = 10.32DD262 pKa = 3.67GEE264 pKa = 4.23IFAVLEE270 pKa = 4.27STVMTEE276 pKa = 3.66EE277 pKa = 3.76FLEE280 pKa = 4.71FFSQHH285 pKa = 5.1WLFEE289 pKa = 4.61ADD291 pKa = 3.21RR292 pKa = 11.84LTWSRR297 pKa = 11.84RR298 pKa = 11.84FKK300 pKa = 10.88QSVSRR305 pKa = 11.84YY306 pKa = 8.41GYY308 pKa = 10.8GFNKK312 pKa = 9.52EE313 pKa = 4.08KK314 pKa = 10.87KK315 pKa = 9.45IDD317 pKa = 3.73TKK319 pKa = 10.98FKK321 pKa = 10.65NLKK324 pKa = 9.76KK325 pKa = 10.23INNIQKK331 pKa = 9.94HH332 pKa = 5.56PPLVGRR338 pKa = 11.84GPCQRR343 pKa = 11.84NLFFEE348 pKa = 4.14NTLRR352 pKa = 11.84SMGGSEE358 pKa = 4.08VRR360 pKa = 11.84AIDD363 pKa = 3.83LYY365 pKa = 11.35KK366 pKa = 10.83NYY368 pKa = 10.27GRR370 pKa = 11.84FGGSRR375 pKa = 11.84VDD377 pKa = 3.94SILYY381 pKa = 7.91MKK383 pKa = 10.7NFDD386 pKa = 4.0KK387 pKa = 10.8KK388 pKa = 11.27YY389 pKa = 7.93PTLDD393 pKa = 4.63LDD395 pKa = 3.59WKK397 pKa = 10.84SNKK400 pKa = 8.0EE401 pKa = 3.51WCFEE405 pKa = 3.68IFGKK409 pKa = 10.6VNHH412 pKa = 6.75IKK414 pKa = 10.64LFDD417 pKa = 3.54MEE419 pKa = 4.54YY420 pKa = 10.6CLDD423 pKa = 3.94VPDD426 pKa = 4.72VPSDD430 pKa = 3.46TSPGITYY437 pKa = 10.15KK438 pKa = 10.85RR439 pKa = 11.84LGFIKK444 pKa = 10.38KK445 pKa = 10.01ADD447 pKa = 4.0ALPVCLSHH455 pKa = 7.82LRR457 pKa = 11.84DD458 pKa = 3.51VLKK461 pKa = 10.71KK462 pKa = 9.77IEE464 pKa = 3.98KK465 pKa = 10.22HH466 pKa = 5.19EE467 pKa = 4.16LQTLPEE473 pKa = 4.32IAWCIAGRR481 pKa = 11.84PKK483 pKa = 9.89FTEE486 pKa = 4.05LNKK489 pKa = 10.36AFQKK493 pKa = 10.88VYY495 pKa = 10.67DD496 pKa = 3.92RR497 pKa = 11.84QSVGRR502 pKa = 11.84SIWVSDD508 pKa = 3.61MEE510 pKa = 4.1EE511 pKa = 4.75AILSRR516 pKa = 11.84HH517 pKa = 4.44FTRR520 pKa = 11.84YY521 pKa = 8.27IDD523 pKa = 3.7KK524 pKa = 10.66NITGTPTFHH533 pKa = 7.85KK534 pKa = 10.65IMINFDD540 pKa = 3.25KK541 pKa = 11.43VKK543 pKa = 10.36DD544 pKa = 3.95AKK546 pKa = 10.65HH547 pKa = 4.89LQNWVEE553 pKa = 3.92QYY555 pKa = 11.46DD556 pKa = 4.08YY557 pKa = 10.66IIEE560 pKa = 4.4ADD562 pKa = 3.55YY563 pKa = 11.62SKK565 pKa = 10.99FDD567 pKa = 3.57SSVPACVVRR576 pKa = 11.84KK577 pKa = 9.74AFQIFKK583 pKa = 10.04EE584 pKa = 4.17LFVSFDD590 pKa = 3.07HH591 pKa = 6.12TTATVYY597 pKa = 11.0DD598 pKa = 3.9FLIEE602 pKa = 4.03NFLNSKK608 pKa = 10.18VDD610 pKa = 3.79LSDD613 pKa = 3.63GRR615 pKa = 11.84LLQKK619 pKa = 10.77FGGVPSGSGFTSIIGSICNYY639 pKa = 10.04IMLDD643 pKa = 3.41TCFMKK648 pKa = 10.21MGLDD652 pKa = 3.48KK653 pKa = 11.16NRR655 pKa = 11.84WSAIVYY661 pKa = 10.6GDD663 pKa = 4.04DD664 pKa = 3.76CLIGIKK670 pKa = 10.25KK671 pKa = 9.35PLHH674 pKa = 5.47KK675 pKa = 10.45HH676 pKa = 5.21EE677 pKa = 5.7DD678 pKa = 3.45SAFKK682 pKa = 10.21IRR684 pKa = 11.84KK685 pKa = 9.0LMLKK689 pKa = 10.26FMQLLFDD696 pKa = 3.71ITLDD700 pKa = 3.78PKK702 pKa = 10.0DD703 pKa = 3.54TKK705 pKa = 10.68LVCEE709 pKa = 4.85KK710 pKa = 9.5YY711 pKa = 10.14VSVIIPEE718 pKa = 4.05YY719 pKa = 11.15DD720 pKa = 3.39EE721 pKa = 5.15DD722 pKa = 4.34TTNGTSLLKK731 pKa = 10.05PSKK734 pKa = 8.66ITRR737 pKa = 11.84IEE739 pKa = 4.07KK740 pKa = 10.74EE741 pKa = 3.62PDD743 pKa = 2.95TFLLSNSSSHH753 pKa = 5.68RR754 pKa = 11.84WWYY757 pKa = 10.93SFEE760 pKa = 4.23KK761 pKa = 8.55TWKK764 pKa = 9.59FLGYY768 pKa = 10.69SLTCDD773 pKa = 3.01GRR775 pKa = 11.84LIRR778 pKa = 11.84PTRR781 pKa = 11.84DD782 pKa = 2.81VLARR786 pKa = 11.84IYY788 pKa = 10.94NPEE791 pKa = 4.13KK792 pKa = 10.17PVKK795 pKa = 10.22SWDD798 pKa = 3.37EE799 pKa = 3.98HH800 pKa = 4.38VTLLKK805 pKa = 9.61MAYY808 pKa = 10.11LEE810 pKa = 4.33NYY812 pKa = 8.58EE813 pKa = 4.23NAHH816 pKa = 4.84TRR818 pKa = 11.84NRR820 pKa = 11.84IYY822 pKa = 10.41HH823 pKa = 5.81YY824 pKa = 11.19LMDD827 pKa = 4.38AWWVIHH833 pKa = 6.27HH834 pKa = 6.84GCPQRR839 pKa = 11.84ALPDD843 pKa = 3.0VDD845 pKa = 3.64YY846 pKa = 9.95TVSKK850 pKa = 10.31GRR852 pKa = 11.84CFHH855 pKa = 7.43RR856 pKa = 11.84YY857 pKa = 5.75TNHH860 pKa = 5.42YY861 pKa = 9.12VHH863 pKa = 6.76LRR865 pKa = 11.84WEE867 pKa = 4.13PSLQGFHH874 pKa = 6.82NFFDD878 pKa = 3.91NFDD881 pKa = 4.01FNMLKK886 pKa = 10.5LHH888 pKa = 6.24IQVSLNEE895 pKa = 4.35FYY897 pKa = 10.96YY898 pKa = 11.0SRR900 pKa = 11.84VKK902 pKa = 10.51RR903 pKa = 11.84GRR905 pKa = 11.84LSSSLFDD912 pKa = 3.2MSMRR916 pKa = 11.84PAHH919 pKa = 5.3YY920 pKa = 10.02HH921 pKa = 4.98SVAYY925 pKa = 10.09LCKK928 pKa = 9.79KK929 pKa = 10.4LGMVTPQVTTAYY941 pKa = 8.56TYY943 pKa = 10.25SKK945 pKa = 10.25EE946 pKa = 4.12WFHH949 pKa = 6.31SHH951 pKa = 7.32PGMLKK956 pKa = 10.24NIKK959 pKa = 9.7KK960 pKa = 9.63VLRR963 pKa = 11.84KK964 pKa = 9.87RR965 pKa = 11.84MQFYY969 pKa = 10.16HH970 pKa = 6.46QDD972 pKa = 3.18GGRR975 pKa = 11.84KK976 pKa = 9.07KK977 pKa = 10.22FKK979 pKa = 10.91KK980 pKa = 9.45MLQNFKK986 pKa = 10.65KK987 pKa = 10.12IRR989 pKa = 11.84QEE991 pKa = 4.21FSFLYY996 pKa = 9.27PNKK999 pKa = 9.7PFTIHH1004 pKa = 6.33NPDD1007 pKa = 3.94IIAALL1012 pKa = 3.68
MM1 pKa = 7.52EE2 pKa = 6.26KK3 pKa = 10.3DD4 pKa = 3.74GKK6 pKa = 10.83SSTWSSFSTALSEE19 pKa = 4.18IQKK22 pKa = 9.92EE23 pKa = 4.16RR24 pKa = 11.84VPGLEE29 pKa = 4.2EE30 pKa = 3.61KK31 pKa = 10.3TSIDD35 pKa = 3.65FEE37 pKa = 4.86VDD39 pKa = 2.78GKK41 pKa = 10.88RR42 pKa = 11.84YY43 pKa = 10.44SLVSSRR49 pKa = 11.84KK50 pKa = 7.12PTDD53 pKa = 3.19RR54 pKa = 11.84EE55 pKa = 4.14VEE57 pKa = 4.27LKK59 pKa = 10.85SLIPSAILTEE69 pKa = 3.86QLLSNEE75 pKa = 4.62HH76 pKa = 5.66IGKK79 pKa = 9.16YY80 pKa = 7.77FEE82 pKa = 5.29RR83 pKa = 11.84IRR85 pKa = 11.84KK86 pKa = 8.68ALIVKK91 pKa = 7.61NTQADD96 pKa = 4.03LVEE99 pKa = 4.58LLNSFTTVLTNIHH112 pKa = 5.73QVSLYY117 pKa = 10.07IEE119 pKa = 4.22HH120 pKa = 7.08HH121 pKa = 7.16PDD123 pKa = 2.76TTAVAKK129 pKa = 10.8SFEE132 pKa = 4.15NYY134 pKa = 9.55KK135 pKa = 10.5LKK137 pKa = 10.72KK138 pKa = 10.23DD139 pKa = 3.6LSKK142 pKa = 10.84WKK144 pKa = 10.6LDD146 pKa = 3.5MLNSLSTVDD155 pKa = 3.69GRR157 pKa = 11.84VQIHH161 pKa = 5.45VNEE164 pKa = 4.04ILAMFHH170 pKa = 5.83GVKK173 pKa = 10.35NAAQKK178 pKa = 9.8DD179 pKa = 3.73AQMKK183 pKa = 8.82EE184 pKa = 3.74EE185 pKa = 4.17KK186 pKa = 10.08ARR188 pKa = 11.84KK189 pKa = 9.1LMKK192 pKa = 10.26QQVEE196 pKa = 4.02LARR199 pKa = 11.84QEE201 pKa = 3.82FEE203 pKa = 3.98EE204 pKa = 4.77RR205 pKa = 11.84IKK207 pKa = 10.65HH208 pKa = 4.57VRR210 pKa = 11.84PFQGLMLLTGSFKK223 pKa = 10.06EE224 pKa = 4.09QSDD227 pKa = 3.98RR228 pKa = 11.84VHH230 pKa = 6.51EE231 pKa = 4.2MARR234 pKa = 11.84QEE236 pKa = 4.26WMSLPDD242 pKa = 3.62TEE244 pKa = 5.47KK245 pKa = 11.18SSDD248 pKa = 3.25YY249 pKa = 11.02DD250 pKa = 2.99AWAQAWWATRR260 pKa = 11.84KK261 pKa = 10.32DD262 pKa = 3.67GEE264 pKa = 4.23IFAVLEE270 pKa = 4.27STVMTEE276 pKa = 3.66EE277 pKa = 3.76FLEE280 pKa = 4.71FFSQHH285 pKa = 5.1WLFEE289 pKa = 4.61ADD291 pKa = 3.21RR292 pKa = 11.84LTWSRR297 pKa = 11.84RR298 pKa = 11.84FKK300 pKa = 10.88QSVSRR305 pKa = 11.84YY306 pKa = 8.41GYY308 pKa = 10.8GFNKK312 pKa = 9.52EE313 pKa = 4.08KK314 pKa = 10.87KK315 pKa = 9.45IDD317 pKa = 3.73TKK319 pKa = 10.98FKK321 pKa = 10.65NLKK324 pKa = 9.76KK325 pKa = 10.23INNIQKK331 pKa = 9.94HH332 pKa = 5.56PPLVGRR338 pKa = 11.84GPCQRR343 pKa = 11.84NLFFEE348 pKa = 4.14NTLRR352 pKa = 11.84SMGGSEE358 pKa = 4.08VRR360 pKa = 11.84AIDD363 pKa = 3.83LYY365 pKa = 11.35KK366 pKa = 10.83NYY368 pKa = 10.27GRR370 pKa = 11.84FGGSRR375 pKa = 11.84VDD377 pKa = 3.94SILYY381 pKa = 7.91MKK383 pKa = 10.7NFDD386 pKa = 4.0KK387 pKa = 10.8KK388 pKa = 11.27YY389 pKa = 7.93PTLDD393 pKa = 4.63LDD395 pKa = 3.59WKK397 pKa = 10.84SNKK400 pKa = 8.0EE401 pKa = 3.51WCFEE405 pKa = 3.68IFGKK409 pKa = 10.6VNHH412 pKa = 6.75IKK414 pKa = 10.64LFDD417 pKa = 3.54MEE419 pKa = 4.54YY420 pKa = 10.6CLDD423 pKa = 3.94VPDD426 pKa = 4.72VPSDD430 pKa = 3.46TSPGITYY437 pKa = 10.15KK438 pKa = 10.85RR439 pKa = 11.84LGFIKK444 pKa = 10.38KK445 pKa = 10.01ADD447 pKa = 4.0ALPVCLSHH455 pKa = 7.82LRR457 pKa = 11.84DD458 pKa = 3.51VLKK461 pKa = 10.71KK462 pKa = 9.77IEE464 pKa = 3.98KK465 pKa = 10.22HH466 pKa = 5.19EE467 pKa = 4.16LQTLPEE473 pKa = 4.32IAWCIAGRR481 pKa = 11.84PKK483 pKa = 9.89FTEE486 pKa = 4.05LNKK489 pKa = 10.36AFQKK493 pKa = 10.88VYY495 pKa = 10.67DD496 pKa = 3.92RR497 pKa = 11.84QSVGRR502 pKa = 11.84SIWVSDD508 pKa = 3.61MEE510 pKa = 4.1EE511 pKa = 4.75AILSRR516 pKa = 11.84HH517 pKa = 4.44FTRR520 pKa = 11.84YY521 pKa = 8.27IDD523 pKa = 3.7KK524 pKa = 10.66NITGTPTFHH533 pKa = 7.85KK534 pKa = 10.65IMINFDD540 pKa = 3.25KK541 pKa = 11.43VKK543 pKa = 10.36DD544 pKa = 3.95AKK546 pKa = 10.65HH547 pKa = 4.89LQNWVEE553 pKa = 3.92QYY555 pKa = 11.46DD556 pKa = 4.08YY557 pKa = 10.66IIEE560 pKa = 4.4ADD562 pKa = 3.55YY563 pKa = 11.62SKK565 pKa = 10.99FDD567 pKa = 3.57SSVPACVVRR576 pKa = 11.84KK577 pKa = 9.74AFQIFKK583 pKa = 10.04EE584 pKa = 4.17LFVSFDD590 pKa = 3.07HH591 pKa = 6.12TTATVYY597 pKa = 11.0DD598 pKa = 3.9FLIEE602 pKa = 4.03NFLNSKK608 pKa = 10.18VDD610 pKa = 3.79LSDD613 pKa = 3.63GRR615 pKa = 11.84LLQKK619 pKa = 10.77FGGVPSGSGFTSIIGSICNYY639 pKa = 10.04IMLDD643 pKa = 3.41TCFMKK648 pKa = 10.21MGLDD652 pKa = 3.48KK653 pKa = 11.16NRR655 pKa = 11.84WSAIVYY661 pKa = 10.6GDD663 pKa = 4.04DD664 pKa = 3.76CLIGIKK670 pKa = 10.25KK671 pKa = 9.35PLHH674 pKa = 5.47KK675 pKa = 10.45HH676 pKa = 5.21EE677 pKa = 5.7DD678 pKa = 3.45SAFKK682 pKa = 10.21IRR684 pKa = 11.84KK685 pKa = 9.0LMLKK689 pKa = 10.26FMQLLFDD696 pKa = 3.71ITLDD700 pKa = 3.78PKK702 pKa = 10.0DD703 pKa = 3.54TKK705 pKa = 10.68LVCEE709 pKa = 4.85KK710 pKa = 9.5YY711 pKa = 10.14VSVIIPEE718 pKa = 4.05YY719 pKa = 11.15DD720 pKa = 3.39EE721 pKa = 5.15DD722 pKa = 4.34TTNGTSLLKK731 pKa = 10.05PSKK734 pKa = 8.66ITRR737 pKa = 11.84IEE739 pKa = 4.07KK740 pKa = 10.74EE741 pKa = 3.62PDD743 pKa = 2.95TFLLSNSSSHH753 pKa = 5.68RR754 pKa = 11.84WWYY757 pKa = 10.93SFEE760 pKa = 4.23KK761 pKa = 8.55TWKK764 pKa = 9.59FLGYY768 pKa = 10.69SLTCDD773 pKa = 3.01GRR775 pKa = 11.84LIRR778 pKa = 11.84PTRR781 pKa = 11.84DD782 pKa = 2.81VLARR786 pKa = 11.84IYY788 pKa = 10.94NPEE791 pKa = 4.13KK792 pKa = 10.17PVKK795 pKa = 10.22SWDD798 pKa = 3.37EE799 pKa = 3.98HH800 pKa = 4.38VTLLKK805 pKa = 9.61MAYY808 pKa = 10.11LEE810 pKa = 4.33NYY812 pKa = 8.58EE813 pKa = 4.23NAHH816 pKa = 4.84TRR818 pKa = 11.84NRR820 pKa = 11.84IYY822 pKa = 10.41HH823 pKa = 5.81YY824 pKa = 11.19LMDD827 pKa = 4.38AWWVIHH833 pKa = 6.27HH834 pKa = 6.84GCPQRR839 pKa = 11.84ALPDD843 pKa = 3.0VDD845 pKa = 3.64YY846 pKa = 9.95TVSKK850 pKa = 10.31GRR852 pKa = 11.84CFHH855 pKa = 7.43RR856 pKa = 11.84YY857 pKa = 5.75TNHH860 pKa = 5.42YY861 pKa = 9.12VHH863 pKa = 6.76LRR865 pKa = 11.84WEE867 pKa = 4.13PSLQGFHH874 pKa = 6.82NFFDD878 pKa = 3.91NFDD881 pKa = 4.01FNMLKK886 pKa = 10.5LHH888 pKa = 6.24IQVSLNEE895 pKa = 4.35FYY897 pKa = 10.96YY898 pKa = 11.0SRR900 pKa = 11.84VKK902 pKa = 10.51RR903 pKa = 11.84GRR905 pKa = 11.84LSSSLFDD912 pKa = 3.2MSMRR916 pKa = 11.84PAHH919 pKa = 5.3YY920 pKa = 10.02HH921 pKa = 4.98SVAYY925 pKa = 10.09LCKK928 pKa = 9.79KK929 pKa = 10.4LGMVTPQVTTAYY941 pKa = 8.56TYY943 pKa = 10.25SKK945 pKa = 10.25EE946 pKa = 4.12WFHH949 pKa = 6.31SHH951 pKa = 7.32PGMLKK956 pKa = 10.24NIKK959 pKa = 9.7KK960 pKa = 9.63VLRR963 pKa = 11.84KK964 pKa = 9.87RR965 pKa = 11.84MQFYY969 pKa = 10.16HH970 pKa = 6.46QDD972 pKa = 3.18GGRR975 pKa = 11.84KK976 pKa = 9.07KK977 pKa = 10.22FKK979 pKa = 10.91KK980 pKa = 9.45MLQNFKK986 pKa = 10.65KK987 pKa = 10.12IRR989 pKa = 11.84QEE991 pKa = 4.21FSFLYY996 pKa = 9.27PNKK999 pKa = 9.7PFTIHH1004 pKa = 6.33NPDD1007 pKa = 3.94IIAALL1012 pKa = 3.68
Molecular weight: 118.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1302 |
290 |
1012 |
651.0 |
76.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.685 ± 0.732 | 1.075 ± 0.517 |
5.914 ± 0.357 | 7.066 ± 1.577 |
5.76 ± 0.449 | 4.147 ± 0.336 |
3.456 ± 0.17 | 5.453 ± 0.467 |
9.524 ± 0.269 | 9.37 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.842 ± 0.126 | 3.763 ± 0.483 |
3.226 ± 0.556 | 3.763 ± 0.678 |
5.376 ± 0.098 | 7.757 ± 0.581 |
5.376 ± 0.234 | 5.684 ± 0.252 |
2.227 ± 0.09 | 3.533 ± 0.87 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
