
Mycobacterium sp. Root135
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium; unclassified Mycobacterium
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
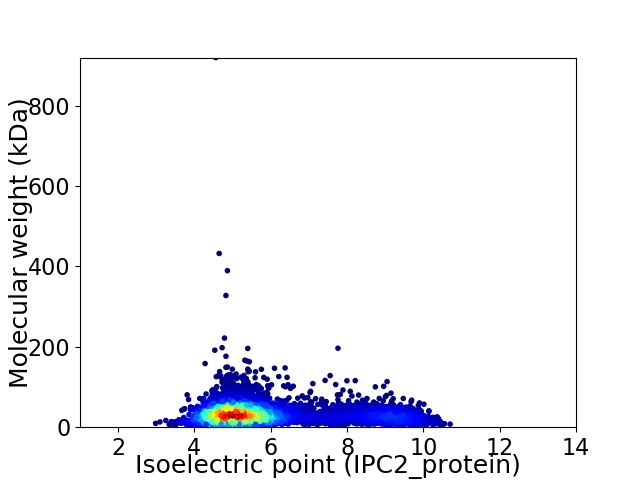
Virtual 2D-PAGE plot for 5995 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0T1WGB6|A0A0T1WGB6_9MYCO Uncharacterized protein OS=Mycobacterium sp. Root135 OX=1736457 GN=ASD37_09440 PE=4 SV=1
MM1 pKa = 6.86PTKK4 pKa = 10.44LAAAFLAAGVISTGALVAPDD24 pKa = 4.35KK25 pKa = 11.44VPLPVVSADD34 pKa = 3.31VANASVITDD43 pKa = 3.69ALYY46 pKa = 10.91RR47 pKa = 11.84LGDD50 pKa = 3.85VVNGAAYY57 pKa = 9.84GYY59 pKa = 10.71AITQDD64 pKa = 3.84AGSSLPFDD72 pKa = 3.53VATAFTIAAQNPTLAPSLLSWLVNRR97 pKa = 11.84YY98 pKa = 9.32ANPSYY103 pKa = 11.07DD104 pKa = 3.13YY105 pKa = 10.61GYY107 pKa = 9.41PSEE110 pKa = 4.88SGYY113 pKa = 8.67FTYY116 pKa = 8.49PTYY119 pKa = 10.47FRR121 pKa = 11.84EE122 pKa = 4.04YY123 pKa = 9.1SLEE126 pKa = 4.32VIAGALPFPLGPAGANPGLINSAANAIGQAVGGFLSGVLPNPAGGLAATDD176 pKa = 4.13AFWATDD182 pKa = 2.97IGKK185 pKa = 8.71TIVAANLAVTAPVWAFYY202 pKa = 9.71STAFYY207 pKa = 10.63LGYY210 pKa = 10.69LPADD214 pKa = 3.92LEE216 pKa = 4.55ATFEE220 pKa = 4.29SAIQNPTEE228 pKa = 4.14IPGLLSNLVYY238 pKa = 10.92DD239 pKa = 4.91LVASDD244 pKa = 4.67GLLGSLIQDD253 pKa = 3.57LSAPLRR259 pKa = 11.84ALPGPIGVLAQNVVASLNAGIDD281 pKa = 3.54NLLSVLPAPIEE292 pKa = 3.95PTPFPSATSNVNRR305 pKa = 11.84VADD308 pKa = 4.41TSAEE312 pKa = 4.03RR313 pKa = 11.84QVTSIPDD320 pKa = 3.46PSITMDD326 pKa = 3.65NVVTLNTPVKK336 pKa = 10.11LDD338 pKa = 3.56NPVAPSSDD346 pKa = 3.78SPPPNAGGTTTGLVKK361 pKa = 10.27TNDD364 pKa = 3.46SGPQLNVVRR373 pKa = 11.84DD374 pKa = 4.1SVKK377 pKa = 9.38VTPGDD382 pKa = 3.42TFAGVTTGTGTGTGTEE398 pKa = 4.41TEE400 pKa = 4.31SATTVDD406 pKa = 3.87EE407 pKa = 4.59TVSGATTASPAAANDD422 pKa = 3.92PSDD425 pKa = 3.38AAAGEE430 pKa = 4.31NVSPSSGAGDD440 pKa = 3.73PGAAAA445 pKa = 4.58
MM1 pKa = 6.86PTKK4 pKa = 10.44LAAAFLAAGVISTGALVAPDD24 pKa = 4.35KK25 pKa = 11.44VPLPVVSADD34 pKa = 3.31VANASVITDD43 pKa = 3.69ALYY46 pKa = 10.91RR47 pKa = 11.84LGDD50 pKa = 3.85VVNGAAYY57 pKa = 9.84GYY59 pKa = 10.71AITQDD64 pKa = 3.84AGSSLPFDD72 pKa = 3.53VATAFTIAAQNPTLAPSLLSWLVNRR97 pKa = 11.84YY98 pKa = 9.32ANPSYY103 pKa = 11.07DD104 pKa = 3.13YY105 pKa = 10.61GYY107 pKa = 9.41PSEE110 pKa = 4.88SGYY113 pKa = 8.67FTYY116 pKa = 8.49PTYY119 pKa = 10.47FRR121 pKa = 11.84EE122 pKa = 4.04YY123 pKa = 9.1SLEE126 pKa = 4.32VIAGALPFPLGPAGANPGLINSAANAIGQAVGGFLSGVLPNPAGGLAATDD176 pKa = 4.13AFWATDD182 pKa = 2.97IGKK185 pKa = 8.71TIVAANLAVTAPVWAFYY202 pKa = 9.71STAFYY207 pKa = 10.63LGYY210 pKa = 10.69LPADD214 pKa = 3.92LEE216 pKa = 4.55ATFEE220 pKa = 4.29SAIQNPTEE228 pKa = 4.14IPGLLSNLVYY238 pKa = 10.92DD239 pKa = 4.91LVASDD244 pKa = 4.67GLLGSLIQDD253 pKa = 3.57LSAPLRR259 pKa = 11.84ALPGPIGVLAQNVVASLNAGIDD281 pKa = 3.54NLLSVLPAPIEE292 pKa = 3.95PTPFPSATSNVNRR305 pKa = 11.84VADD308 pKa = 4.41TSAEE312 pKa = 4.03RR313 pKa = 11.84QVTSIPDD320 pKa = 3.46PSITMDD326 pKa = 3.65NVVTLNTPVKK336 pKa = 10.11LDD338 pKa = 3.56NPVAPSSDD346 pKa = 3.78SPPPNAGGTTTGLVKK361 pKa = 10.27TNDD364 pKa = 3.46SGPQLNVVRR373 pKa = 11.84DD374 pKa = 4.1SVKK377 pKa = 9.38VTPGDD382 pKa = 3.42TFAGVTTGTGTGTGTEE398 pKa = 4.41TEE400 pKa = 4.31SATTVDD406 pKa = 3.87EE407 pKa = 4.59TVSGATTASPAAANDD422 pKa = 3.92PSDD425 pKa = 3.38AAAGEE430 pKa = 4.31NVSPSSGAGDD440 pKa = 3.73PGAAAA445 pKa = 4.58
Molecular weight: 44.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0T1WG68|A0A0T1WG68_9MYCO Aminotransferase class V OS=Mycobacterium sp. Root135 OX=1736457 GN=ASD37_09005 PE=4 SV=1
MM1 pKa = 7.41SRR3 pKa = 11.84PEE5 pKa = 3.94GQPARR10 pKa = 11.84GLRR13 pKa = 11.84ILPSATTVLAICAGLTSIKK32 pKa = 10.15FALDD36 pKa = 3.33EE37 pKa = 4.66KK38 pKa = 10.52PWISLALIGAAAILDD53 pKa = 4.84GIDD56 pKa = 3.22GGIARR61 pKa = 11.84ALNAQSRR68 pKa = 11.84MGAEE72 pKa = 3.94IDD74 pKa = 3.88SLADD78 pKa = 3.28AVNFGVAPALVVYY91 pKa = 8.04LTLLPNSQLGWIFVLLYY108 pKa = 10.08AVCVVLRR115 pKa = 11.84LARR118 pKa = 11.84FNALLDD124 pKa = 3.85DD125 pKa = 4.12HH126 pKa = 6.93TKK128 pKa = 9.84PAYY131 pKa = 9.06TRR133 pKa = 11.84EE134 pKa = 4.06FFTGVPAPCGAVGALGPLVAFMQFGHH160 pKa = 6.17GWWTSHH166 pKa = 5.63WFVCAWLAANAALLVSRR183 pKa = 11.84VPTLALKK190 pKa = 10.53AISVPEE196 pKa = 3.78NAAPILLILVAAAAAGLLLFPYY218 pKa = 10.43VLVLLIIVGYY228 pKa = 9.07VCMMPFTIRR237 pKa = 11.84SQRR240 pKa = 11.84WVAARR245 pKa = 11.84PEE247 pKa = 3.94AWDD250 pKa = 3.56AKK252 pKa = 9.06PGQRR256 pKa = 11.84RR257 pKa = 11.84AVRR260 pKa = 11.84RR261 pKa = 11.84AARR264 pKa = 11.84RR265 pKa = 11.84AAPNRR270 pKa = 11.84RR271 pKa = 11.84SVARR275 pKa = 11.84LGLRR279 pKa = 11.84KK280 pKa = 9.59PGSRR284 pKa = 11.84GG285 pKa = 3.17
MM1 pKa = 7.41SRR3 pKa = 11.84PEE5 pKa = 3.94GQPARR10 pKa = 11.84GLRR13 pKa = 11.84ILPSATTVLAICAGLTSIKK32 pKa = 10.15FALDD36 pKa = 3.33EE37 pKa = 4.66KK38 pKa = 10.52PWISLALIGAAAILDD53 pKa = 4.84GIDD56 pKa = 3.22GGIARR61 pKa = 11.84ALNAQSRR68 pKa = 11.84MGAEE72 pKa = 3.94IDD74 pKa = 3.88SLADD78 pKa = 3.28AVNFGVAPALVVYY91 pKa = 8.04LTLLPNSQLGWIFVLLYY108 pKa = 10.08AVCVVLRR115 pKa = 11.84LARR118 pKa = 11.84FNALLDD124 pKa = 3.85DD125 pKa = 4.12HH126 pKa = 6.93TKK128 pKa = 9.84PAYY131 pKa = 9.06TRR133 pKa = 11.84EE134 pKa = 4.06FFTGVPAPCGAVGALGPLVAFMQFGHH160 pKa = 6.17GWWTSHH166 pKa = 5.63WFVCAWLAANAALLVSRR183 pKa = 11.84VPTLALKK190 pKa = 10.53AISVPEE196 pKa = 3.78NAAPILLILVAAAAAGLLLFPYY218 pKa = 10.43VLVLLIIVGYY228 pKa = 9.07VCMMPFTIRR237 pKa = 11.84SQRR240 pKa = 11.84WVAARR245 pKa = 11.84PEE247 pKa = 3.94AWDD250 pKa = 3.56AKK252 pKa = 9.06PGQRR256 pKa = 11.84RR257 pKa = 11.84AVRR260 pKa = 11.84RR261 pKa = 11.84AARR264 pKa = 11.84RR265 pKa = 11.84AAPNRR270 pKa = 11.84RR271 pKa = 11.84SVARR275 pKa = 11.84LGLRR279 pKa = 11.84KK280 pKa = 9.59PGSRR284 pKa = 11.84GG285 pKa = 3.17
Molecular weight: 30.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1952207 |
31 |
8621 |
325.6 |
34.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.946 ± 0.04 | 0.764 ± 0.009 |
6.523 ± 0.027 | 5.106 ± 0.025 |
3.116 ± 0.019 | 8.988 ± 0.033 |
2.149 ± 0.016 | 4.01 ± 0.02 |
2.209 ± 0.019 | 9.823 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.058 ± 0.013 | 2.223 ± 0.017 |
5.647 ± 0.029 | 2.816 ± 0.016 |
6.906 ± 0.031 | 5.547 ± 0.018 |
6.271 ± 0.023 | 9.321 ± 0.032 |
1.476 ± 0.015 | 2.099 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
