
Singapore grouper iridovirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Iridoviridae; Alphairidovirinae; Ranavirus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
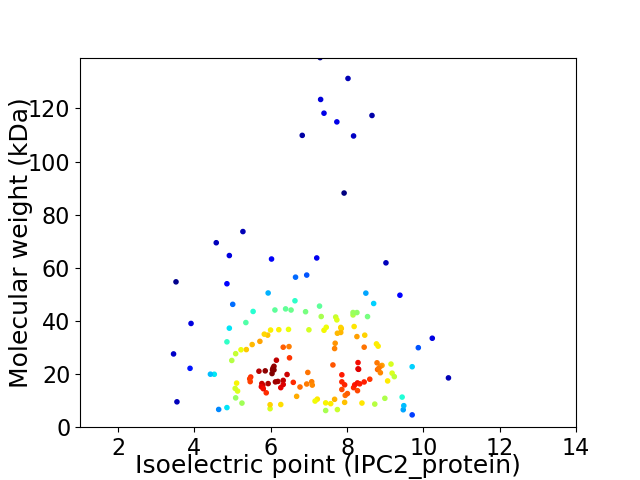
Virtual 2D-PAGE plot for 162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
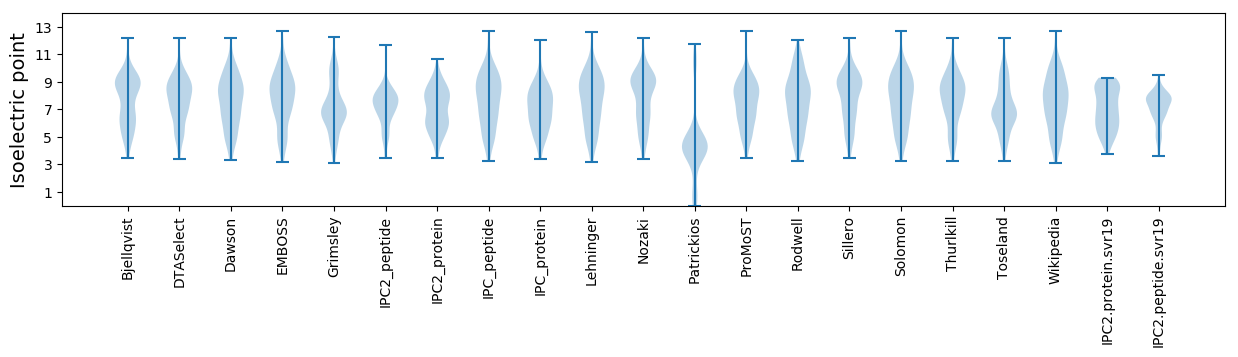
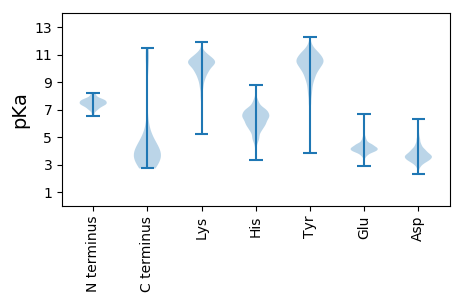
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5YFK3|Q5YFK3_9VIRU IlGF domain-containing protein OS=Singapore grouper iridovirus OX=262968 GN=ORF062R PE=4 SV=1
MM1 pKa = 7.44GSGVVSGDD9 pKa = 3.4SCVAAGDD16 pKa = 4.01DD17 pKa = 3.59VGDD20 pKa = 3.98FSVVSSYY27 pKa = 10.72IPEE30 pKa = 4.55GSWVAWGEE38 pKa = 4.05EE39 pKa = 4.45GSWVAWGEE47 pKa = 4.05EE48 pKa = 4.45GSWVAWGEE56 pKa = 4.05EE57 pKa = 4.45GSWVAWGEE65 pKa = 4.05EE66 pKa = 4.45GSWVAWGEE74 pKa = 4.05EE75 pKa = 4.45GSWVAWGEE83 pKa = 4.05EE84 pKa = 4.45GSWVAWGEE92 pKa = 4.05EE93 pKa = 4.45GSWVAWGEE101 pKa = 4.05EE102 pKa = 4.45GSWVAWGEE110 pKa = 4.05EE111 pKa = 4.45GSWVAWGEE119 pKa = 3.91EE120 pKa = 4.37GSWVISSVIPKK131 pKa = 7.74DD132 pKa = 3.76TPISGATVEE141 pKa = 4.35GATVEE146 pKa = 4.25GATVDD151 pKa = 4.28FLGLQYY157 pKa = 10.66RR158 pKa = 11.84SNSFSSHH165 pKa = 5.98SFLQHH170 pKa = 5.56ASARR174 pKa = 11.84PTLRR178 pKa = 11.84VLLPLLGGVYY188 pKa = 9.8TPLSPQCSVSASINSPPHH206 pKa = 5.02TCSS209 pKa = 2.98
MM1 pKa = 7.44GSGVVSGDD9 pKa = 3.4SCVAAGDD16 pKa = 4.01DD17 pKa = 3.59VGDD20 pKa = 3.98FSVVSSYY27 pKa = 10.72IPEE30 pKa = 4.55GSWVAWGEE38 pKa = 4.05EE39 pKa = 4.45GSWVAWGEE47 pKa = 4.05EE48 pKa = 4.45GSWVAWGEE56 pKa = 4.05EE57 pKa = 4.45GSWVAWGEE65 pKa = 4.05EE66 pKa = 4.45GSWVAWGEE74 pKa = 4.05EE75 pKa = 4.45GSWVAWGEE83 pKa = 4.05EE84 pKa = 4.45GSWVAWGEE92 pKa = 4.05EE93 pKa = 4.45GSWVAWGEE101 pKa = 4.05EE102 pKa = 4.45GSWVAWGEE110 pKa = 4.05EE111 pKa = 4.45GSWVAWGEE119 pKa = 3.91EE120 pKa = 4.37GSWVISSVIPKK131 pKa = 7.74DD132 pKa = 3.76TPISGATVEE141 pKa = 4.35GATVEE146 pKa = 4.25GATVDD151 pKa = 4.28FLGLQYY157 pKa = 10.66RR158 pKa = 11.84SNSFSSHH165 pKa = 5.98SFLQHH170 pKa = 5.56ASARR174 pKa = 11.84PTLRR178 pKa = 11.84VLLPLLGGVYY188 pKa = 9.8TPLSPQCSVSASINSPPHH206 pKa = 5.02TCSS209 pKa = 2.98
Molecular weight: 22.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5YFG1|Q5YFG1_9VIRU DNA-directed RNA polymerase subunit OS=Singapore grouper iridovirus OX=262968 GN=ORF104L PE=3 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84AAQASSAGRR11 pKa = 11.84QSTYY15 pKa = 9.5PAEE18 pKa = 4.51NGLHH22 pKa = 5.69VSHH25 pKa = 7.42LLTINCTLSVNSAAASSPYY44 pKa = 10.57SKK46 pKa = 10.56YY47 pKa = 10.44SNFMIVFKK55 pKa = 10.54IFLVTCPLTILPAVMTASLHH75 pKa = 6.0FKK77 pKa = 10.44LKK79 pKa = 10.7SRR81 pKa = 11.84GSNSSNKK88 pKa = 9.51PKK90 pKa = 10.92GDD92 pKa = 3.42VASAAGPVEE101 pKa = 4.25ASHH104 pKa = 6.91PSIPNASNKK113 pKa = 7.5QHH115 pKa = 6.42ALSSRR120 pKa = 11.84GLGSRR125 pKa = 11.84FILMRR130 pKa = 11.84CAPSTASCTNTTSADD145 pKa = 3.58GPRR148 pKa = 11.84VLIMTLRR155 pKa = 11.84GAINVNVSAMAWGPCGKK172 pKa = 9.84YY173 pKa = 9.81RR174 pKa = 11.84LPSSPSKK181 pKa = 11.38SMLKK185 pKa = 10.22PAVTDD190 pKa = 3.52RR191 pKa = 11.84LSRR194 pKa = 11.84MVFPGSRR201 pKa = 11.84FTLIPSIEE209 pKa = 3.94LRR211 pKa = 11.84CRR213 pKa = 11.84VGCLLRR219 pKa = 11.84ATEE222 pKa = 4.45SPSSKK227 pKa = 9.16TRR229 pKa = 11.84STVIPGSNLGHH240 pKa = 6.36SNSSPRR246 pKa = 11.84SFTRR250 pKa = 11.84PVTWHH255 pKa = 5.82RR256 pKa = 11.84TTTFFRR262 pKa = 11.84AAVLAGCVTLPSSLPHH278 pKa = 5.69SVSMM282 pKa = 5.16
MM1 pKa = 7.75RR2 pKa = 11.84AAQASSAGRR11 pKa = 11.84QSTYY15 pKa = 9.5PAEE18 pKa = 4.51NGLHH22 pKa = 5.69VSHH25 pKa = 7.42LLTINCTLSVNSAAASSPYY44 pKa = 10.57SKK46 pKa = 10.56YY47 pKa = 10.44SNFMIVFKK55 pKa = 10.54IFLVTCPLTILPAVMTASLHH75 pKa = 6.0FKK77 pKa = 10.44LKK79 pKa = 10.7SRR81 pKa = 11.84GSNSSNKK88 pKa = 9.51PKK90 pKa = 10.92GDD92 pKa = 3.42VASAAGPVEE101 pKa = 4.25ASHH104 pKa = 6.91PSIPNASNKK113 pKa = 7.5QHH115 pKa = 6.42ALSSRR120 pKa = 11.84GLGSRR125 pKa = 11.84FILMRR130 pKa = 11.84CAPSTASCTNTTSADD145 pKa = 3.58GPRR148 pKa = 11.84VLIMTLRR155 pKa = 11.84GAINVNVSAMAWGPCGKK172 pKa = 9.84YY173 pKa = 9.81RR174 pKa = 11.84LPSSPSKK181 pKa = 11.38SMLKK185 pKa = 10.22PAVTDD190 pKa = 3.52RR191 pKa = 11.84LSRR194 pKa = 11.84MVFPGSRR201 pKa = 11.84FTLIPSIEE209 pKa = 3.94LRR211 pKa = 11.84CRR213 pKa = 11.84VGCLLRR219 pKa = 11.84ATEE222 pKa = 4.45SPSSKK227 pKa = 9.16TRR229 pKa = 11.84STVIPGSNLGHH240 pKa = 6.36SNSSPRR246 pKa = 11.84SFTRR250 pKa = 11.84PVTWHH255 pKa = 5.82RR256 pKa = 11.84TTTFFRR262 pKa = 11.84AAVLAGCVTLPSSLPHH278 pKa = 5.69SVSMM282 pKa = 5.16
Molecular weight: 29.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
46194 |
41 |
1268 |
285.1 |
31.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.293 ± 0.408 | 2.36 ± 0.15 |
5.059 ± 0.147 | 6.029 ± 0.283 |
3.695 ± 0.18 | 6.932 ± 0.419 |
2.054 ± 0.101 | 4.674 ± 0.164 |
6.053 ± 0.199 | 8.168 ± 0.3 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.782 ± 0.109 | 3.931 ± 0.166 |
6.107 ± 0.38 | 2.99 ± 0.145 |
5.494 ± 0.243 | 7.306 ± 0.356 |
6.291 ± 0.186 | 7.113 ± 0.213 |
1.442 ± 0.129 | 3.228 ± 0.172 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
