
Avon-Heathcote Estuary associated circular virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.61
Get precalculated fractions of proteins
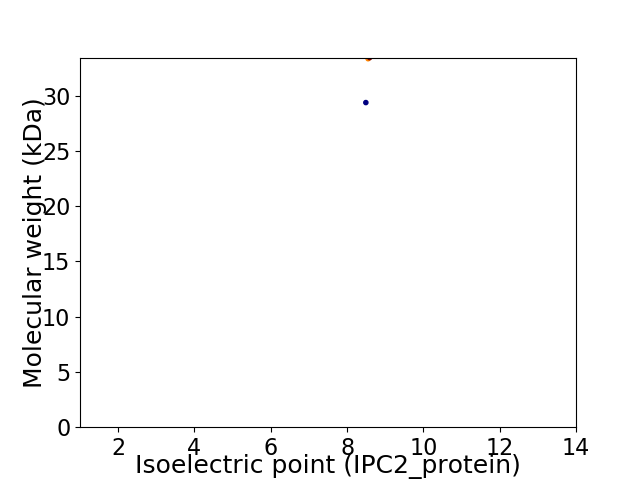
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBG0|A0A0C5IBG0_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 2 OX=1618243 PE=4 SV=1
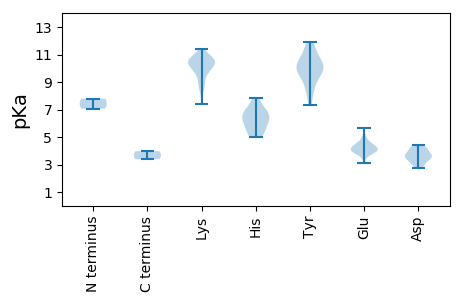
MM1 pKa = 7.8PRR3 pKa = 11.84GQLTKK8 pKa = 10.03TEE10 pKa = 4.06KK11 pKa = 10.51KK12 pKa = 8.66QVKK15 pKa = 10.36NIVQSQLSKK24 pKa = 9.7VTEE27 pKa = 4.44LKK29 pKa = 9.66QHH31 pKa = 4.99VVEE34 pKa = 5.11FDD36 pKa = 3.4YY37 pKa = 11.59DD38 pKa = 3.65PVFRR42 pKa = 11.84LNAGGVFHH50 pKa = 7.08CLSNVQLNTSSPAGMTDD67 pKa = 2.78ATRR70 pKa = 11.84NEE72 pKa = 4.08RR73 pKa = 11.84FGSKK77 pKa = 9.86ISPKK81 pKa = 10.48SITVDD86 pKa = 3.14LMLRR90 pKa = 11.84GSRR93 pKa = 11.84DD94 pKa = 3.5SVGFSGKK101 pKa = 8.11TVDD104 pKa = 4.12EE105 pKa = 4.06IRR107 pKa = 11.84VVLFRR112 pKa = 11.84WKK114 pKa = 11.03NEE116 pKa = 3.63TDD118 pKa = 3.57VVAGGTKK125 pKa = 10.17FPTQGLLWGEE135 pKa = 4.38TSPDD139 pKa = 3.07QYY141 pKa = 11.89NILTKK146 pKa = 10.0PLNVFGDD153 pKa = 3.48KK154 pKa = 10.82DD155 pKa = 3.81FEE157 pKa = 4.28IVEE160 pKa = 4.12DD161 pKa = 4.23RR162 pKa = 11.84KK163 pKa = 10.77IKK165 pKa = 9.76LTGDD169 pKa = 3.31ASQYY173 pKa = 9.38MSGVSLISHH182 pKa = 6.57TNSSQNKK189 pKa = 9.58DD190 pKa = 2.75KK191 pKa = 11.08LLRR194 pKa = 11.84LKK196 pKa = 10.74VPYY199 pKa = 10.08KK200 pKa = 10.83DD201 pKa = 3.13IVKK204 pKa = 8.95TLNFVDD210 pKa = 4.11SQVGTGFHH218 pKa = 5.67TAEE221 pKa = 4.38HH222 pKa = 6.61RR223 pKa = 11.84NSYY226 pKa = 8.23WLYY229 pKa = 9.98IATDD233 pKa = 3.96NNIMGHH239 pKa = 6.01PVTMNLPYY247 pKa = 8.62YY248 pKa = 9.33TLYY251 pKa = 11.3SRR253 pKa = 11.84MNFTDD258 pKa = 3.17IGAA261 pKa = 3.97
MM1 pKa = 7.8PRR3 pKa = 11.84GQLTKK8 pKa = 10.03TEE10 pKa = 4.06KK11 pKa = 10.51KK12 pKa = 8.66QVKK15 pKa = 10.36NIVQSQLSKK24 pKa = 9.7VTEE27 pKa = 4.44LKK29 pKa = 9.66QHH31 pKa = 4.99VVEE34 pKa = 5.11FDD36 pKa = 3.4YY37 pKa = 11.59DD38 pKa = 3.65PVFRR42 pKa = 11.84LNAGGVFHH50 pKa = 7.08CLSNVQLNTSSPAGMTDD67 pKa = 2.78ATRR70 pKa = 11.84NEE72 pKa = 4.08RR73 pKa = 11.84FGSKK77 pKa = 9.86ISPKK81 pKa = 10.48SITVDD86 pKa = 3.14LMLRR90 pKa = 11.84GSRR93 pKa = 11.84DD94 pKa = 3.5SVGFSGKK101 pKa = 8.11TVDD104 pKa = 4.12EE105 pKa = 4.06IRR107 pKa = 11.84VVLFRR112 pKa = 11.84WKK114 pKa = 11.03NEE116 pKa = 3.63TDD118 pKa = 3.57VVAGGTKK125 pKa = 10.17FPTQGLLWGEE135 pKa = 4.38TSPDD139 pKa = 3.07QYY141 pKa = 11.89NILTKK146 pKa = 10.0PLNVFGDD153 pKa = 3.48KK154 pKa = 10.82DD155 pKa = 3.81FEE157 pKa = 4.28IVEE160 pKa = 4.12DD161 pKa = 4.23RR162 pKa = 11.84KK163 pKa = 10.77IKK165 pKa = 9.76LTGDD169 pKa = 3.31ASQYY173 pKa = 9.38MSGVSLISHH182 pKa = 6.57TNSSQNKK189 pKa = 9.58DD190 pKa = 2.75KK191 pKa = 11.08LLRR194 pKa = 11.84LKK196 pKa = 10.74VPYY199 pKa = 10.08KK200 pKa = 10.83DD201 pKa = 3.13IVKK204 pKa = 8.95TLNFVDD210 pKa = 4.11SQVGTGFHH218 pKa = 5.67TAEE221 pKa = 4.38HH222 pKa = 6.61RR223 pKa = 11.84NSYY226 pKa = 8.23WLYY229 pKa = 9.98IATDD233 pKa = 3.96NNIMGHH239 pKa = 6.01PVTMNLPYY247 pKa = 8.62YY248 pKa = 9.33TLYY251 pKa = 11.3SRR253 pKa = 11.84MNFTDD258 pKa = 3.17IGAA261 pKa = 3.97
Molecular weight: 29.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBG0|A0A0C5IBG0_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 2 OX=1618243 PE=4 SV=1
MM1 pKa = 7.06KK2 pKa = 10.62QNTKK6 pKa = 10.55KK7 pKa = 9.86EE8 pKa = 3.75LRR10 pKa = 11.84YY11 pKa = 9.88RR12 pKa = 11.84RR13 pKa = 11.84WCITEE18 pKa = 3.93NNYY21 pKa = 9.27TDD23 pKa = 3.72EE24 pKa = 4.6SIKK27 pKa = 10.5ILSEE31 pKa = 3.92LPTNYY36 pKa = 9.7IIVGKK41 pKa = 10.37EE42 pKa = 3.08IGKK45 pKa = 9.84KK46 pKa = 10.09KK47 pKa = 9.02KK48 pKa = 8.64TPHH51 pKa = 5.18LQIYY55 pKa = 10.43LEE57 pKa = 4.1FVNAKK62 pKa = 8.55TFSAVKK68 pKa = 10.26KK69 pKa = 9.49LLKK72 pKa = 9.7KK73 pKa = 10.64AHH75 pKa = 6.46IEE77 pKa = 4.04VSKK80 pKa = 11.42GNAKK84 pKa = 10.34QNTDD88 pKa = 3.67YY89 pKa = 11.02CSKK92 pKa = 10.48DD93 pKa = 3.02QDD95 pKa = 4.04VILVRR100 pKa = 11.84GEE102 pKa = 3.97PFNQGKK108 pKa = 10.29RR109 pKa = 11.84NDD111 pKa = 3.66LSCIKK116 pKa = 10.38EE117 pKa = 4.83LIQQDD122 pKa = 3.77YY123 pKa = 10.55NIRR126 pKa = 11.84EE127 pKa = 4.12MLEE130 pKa = 3.92EE131 pKa = 4.01EE132 pKa = 4.86SIKK135 pKa = 10.7SLQGLRR141 pKa = 11.84MAEE144 pKa = 3.7NLMKK148 pKa = 10.66YY149 pKa = 8.46VEE151 pKa = 4.58KK152 pKa = 10.4PRR154 pKa = 11.84SKK156 pKa = 10.68APNIIWRR163 pKa = 11.84YY164 pKa = 7.33GASGSGKK171 pKa = 7.41TRR173 pKa = 11.84WVYY176 pKa = 10.97EE177 pKa = 3.69NYY179 pKa = 10.11TEE181 pKa = 4.35VFCPINFKK189 pKa = 9.01WWEE192 pKa = 4.39GYY194 pKa = 9.96DD195 pKa = 3.43GHH197 pKa = 7.8KK198 pKa = 10.7VVLLDD203 pKa = 4.13DD204 pKa = 4.42LRR206 pKa = 11.84GDD208 pKa = 3.46FCKK211 pKa = 10.65YY212 pKa = 10.56HH213 pKa = 6.95EE214 pKa = 4.43FLKK217 pKa = 10.3LTDD220 pKa = 4.02RR221 pKa = 11.84YY222 pKa = 9.73PYY224 pKa = 10.22RR225 pKa = 11.84VEE227 pKa = 4.09CKK229 pKa = 10.25GGSRR233 pKa = 11.84QLLADD238 pKa = 4.4TIIITSPVHH247 pKa = 6.33PKK249 pKa = 10.1EE250 pKa = 4.0VWDD253 pKa = 4.26TIEE256 pKa = 5.68DD257 pKa = 3.64KK258 pKa = 11.06KK259 pKa = 11.15QLLRR263 pKa = 11.84RR264 pKa = 11.84ITKK267 pKa = 9.07IDD269 pKa = 3.64HH270 pKa = 5.67VVEE273 pKa = 4.5NYY275 pKa = 10.18TEE277 pKa = 4.23VKK279 pKa = 10.35RR280 pKa = 11.84GNTT283 pKa = 3.38
MM1 pKa = 7.06KK2 pKa = 10.62QNTKK6 pKa = 10.55KK7 pKa = 9.86EE8 pKa = 3.75LRR10 pKa = 11.84YY11 pKa = 9.88RR12 pKa = 11.84RR13 pKa = 11.84WCITEE18 pKa = 3.93NNYY21 pKa = 9.27TDD23 pKa = 3.72EE24 pKa = 4.6SIKK27 pKa = 10.5ILSEE31 pKa = 3.92LPTNYY36 pKa = 9.7IIVGKK41 pKa = 10.37EE42 pKa = 3.08IGKK45 pKa = 9.84KK46 pKa = 10.09KK47 pKa = 9.02KK48 pKa = 8.64TPHH51 pKa = 5.18LQIYY55 pKa = 10.43LEE57 pKa = 4.1FVNAKK62 pKa = 8.55TFSAVKK68 pKa = 10.26KK69 pKa = 9.49LLKK72 pKa = 9.7KK73 pKa = 10.64AHH75 pKa = 6.46IEE77 pKa = 4.04VSKK80 pKa = 11.42GNAKK84 pKa = 10.34QNTDD88 pKa = 3.67YY89 pKa = 11.02CSKK92 pKa = 10.48DD93 pKa = 3.02QDD95 pKa = 4.04VILVRR100 pKa = 11.84GEE102 pKa = 3.97PFNQGKK108 pKa = 10.29RR109 pKa = 11.84NDD111 pKa = 3.66LSCIKK116 pKa = 10.38EE117 pKa = 4.83LIQQDD122 pKa = 3.77YY123 pKa = 10.55NIRR126 pKa = 11.84EE127 pKa = 4.12MLEE130 pKa = 3.92EE131 pKa = 4.01EE132 pKa = 4.86SIKK135 pKa = 10.7SLQGLRR141 pKa = 11.84MAEE144 pKa = 3.7NLMKK148 pKa = 10.66YY149 pKa = 8.46VEE151 pKa = 4.58KK152 pKa = 10.4PRR154 pKa = 11.84SKK156 pKa = 10.68APNIIWRR163 pKa = 11.84YY164 pKa = 7.33GASGSGKK171 pKa = 7.41TRR173 pKa = 11.84WVYY176 pKa = 10.97EE177 pKa = 3.69NYY179 pKa = 10.11TEE181 pKa = 4.35VFCPINFKK189 pKa = 9.01WWEE192 pKa = 4.39GYY194 pKa = 9.96DD195 pKa = 3.43GHH197 pKa = 7.8KK198 pKa = 10.7VVLLDD203 pKa = 4.13DD204 pKa = 4.42LRR206 pKa = 11.84GDD208 pKa = 3.46FCKK211 pKa = 10.65YY212 pKa = 10.56HH213 pKa = 6.95EE214 pKa = 4.43FLKK217 pKa = 10.3LTDD220 pKa = 4.02RR221 pKa = 11.84YY222 pKa = 9.73PYY224 pKa = 10.22RR225 pKa = 11.84VEE227 pKa = 4.09CKK229 pKa = 10.25GGSRR233 pKa = 11.84QLLADD238 pKa = 4.4TIIITSPVHH247 pKa = 6.33PKK249 pKa = 10.1EE250 pKa = 4.0VWDD253 pKa = 4.26TIEE256 pKa = 5.68DD257 pKa = 3.64KK258 pKa = 11.06KK259 pKa = 11.15QLLRR263 pKa = 11.84RR264 pKa = 11.84ITKK267 pKa = 9.07IDD269 pKa = 3.64HH270 pKa = 5.67VVEE273 pKa = 4.5NYY275 pKa = 10.18TEE277 pKa = 4.23VKK279 pKa = 10.35RR280 pKa = 11.84GNTT283 pKa = 3.38
Molecular weight: 33.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
544 |
261 |
283 |
272.0 |
31.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.941 ± 0.088 | 1.287 ± 0.642 |
5.882 ± 0.448 | 6.25 ± 1.718 |
3.493 ± 0.785 | 6.25 ± 0.731 |
2.206 ± 0.066 | 6.25 ± 1.174 |
9.926 ± 1.608 | 8.272 ± 0.112 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.022 ± 0.469 | 5.882 ± 0.176 |
3.493 ± 0.241 | 3.86 ± 0.252 |
5.331 ± 0.521 | 6.25 ± 1.276 |
6.985 ± 1.025 | 7.353 ± 1.036 |
1.654 ± 0.359 | 4.412 ± 0.684 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
