
Persimmon cryptic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
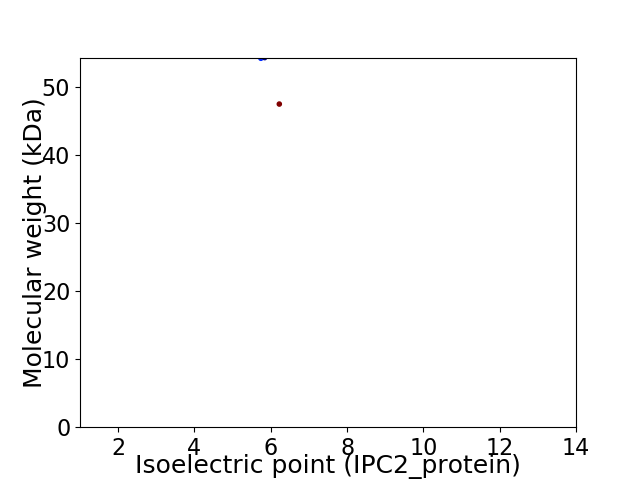
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4EPL9|I4EPL9_9VIRU Coat protein OS=Persimmon cryptic virus OX=1183241 GN=CP PE=4 SV=1
MM1 pKa = 7.9ALRR4 pKa = 11.84SITGYY9 pKa = 10.15EE10 pKa = 3.73FHH12 pKa = 7.74DD13 pKa = 4.06FQSSLEE19 pKa = 4.3LLNQTHH25 pKa = 5.14IHH27 pKa = 5.36IVRR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 4.06SGVTYY37 pKa = 10.15HH38 pKa = 7.71DD39 pKa = 3.92EE40 pKa = 4.1FALCEE45 pKa = 4.03LLVDD49 pKa = 3.6NTRR52 pKa = 11.84LYY54 pKa = 10.15EE55 pKa = 3.97QEE57 pKa = 3.94LEE59 pKa = 4.18GWSRR63 pKa = 11.84SYY65 pKa = 10.01YY66 pKa = 10.1TGEE69 pKa = 3.75QHH71 pKa = 6.39MKK73 pKa = 10.8AILQYY78 pKa = 10.62SLPNTPIQDD87 pKa = 3.17IDD89 pKa = 3.78VGCYY93 pKa = 6.98QQAMTNVQEE102 pKa = 4.56RR103 pKa = 11.84LSSLPIVRR111 pKa = 11.84AFDD114 pKa = 3.43VLTQLDD120 pKa = 3.83QVSFEE125 pKa = 4.25SSSAAGYY132 pKa = 10.44DD133 pKa = 3.42YY134 pKa = 11.25TGAKK138 pKa = 9.43GPKK141 pKa = 9.39NEE143 pKa = 4.37GNHH146 pKa = 4.54EE147 pKa = 3.95RR148 pKa = 11.84AIRR151 pKa = 11.84RR152 pKa = 11.84AKK154 pKa = 9.99AVLWSAIAQDD164 pKa = 4.06GEE166 pKa = 4.64GIEE169 pKa = 4.26HH170 pKa = 6.53VLRR173 pKa = 11.84SSVPDD178 pKa = 3.17VGYY181 pKa = 8.77TRR183 pKa = 11.84TQLTDD188 pKa = 3.48LSEE191 pKa = 4.12KK192 pKa = 9.74TKK194 pKa = 10.6VRR196 pKa = 11.84GVWGRR201 pKa = 11.84AFHH204 pKa = 6.63YY205 pKa = 9.76ILPEE209 pKa = 4.15GTSADD214 pKa = 3.84PLLQAFKK221 pKa = 10.87EE222 pKa = 4.45GGTFYY227 pKa = 10.77HH228 pKa = 7.2IGQDD232 pKa = 3.49PTVSVPYY239 pKa = 10.01ILSDD243 pKa = 3.6TAGKK247 pKa = 9.67CAWLYY252 pKa = 11.14ALDD255 pKa = 3.85WSSFDD260 pKa = 3.26ATVSRR265 pKa = 11.84FEE267 pKa = 3.48IHH269 pKa = 7.07AAFDD273 pKa = 3.93LLKK276 pKa = 10.62QRR278 pKa = 11.84IEE280 pKa = 4.08FPNFEE285 pKa = 4.2TEE287 pKa = 3.56QCYY290 pKa = 9.72EE291 pKa = 3.47ICRR294 pKa = 11.84QLFIHH299 pKa = 6.58KK300 pKa = 9.99KK301 pKa = 8.91IAAPDD306 pKa = 3.68GKK308 pKa = 10.84VYY310 pKa = 9.56WAHH313 pKa = 6.77KK314 pKa = 8.99GIPSGSYY321 pKa = 6.92YY322 pKa = 10.46TSIIGSIINRR332 pKa = 11.84LRR334 pKa = 11.84IEE336 pKa = 4.37YY337 pKa = 9.63IWIKK341 pKa = 10.68LRR343 pKa = 11.84GHH345 pKa = 6.83GPTICYY351 pKa = 7.61TQGDD355 pKa = 4.29DD356 pKa = 4.56SLCGDD361 pKa = 4.47DD362 pKa = 5.6EE363 pKa = 5.59RR364 pKa = 11.84IEE366 pKa = 4.09PEE368 pKa = 4.58RR369 pKa = 11.84IADD372 pKa = 3.46IANPIGWLINPAKK385 pKa = 9.63TATTRR390 pKa = 11.84YY391 pKa = 9.02PEE393 pKa = 4.52YY394 pKa = 9.68ITFLGRR400 pKa = 11.84TCYY403 pKa = 10.48GGLNHH408 pKa = 7.54RR409 pKa = 11.84DD410 pKa = 3.64LIRR413 pKa = 11.84CLRR416 pKa = 11.84LLIYY420 pKa = 10.05PEE422 pKa = 4.19YY423 pKa = 9.12PVPSGAISAYY433 pKa = 8.96RR434 pKa = 11.84ANSIAEE440 pKa = 4.24DD441 pKa = 3.75CGGTSSILNDD451 pKa = 2.94IARR454 pKa = 11.84RR455 pKa = 11.84LTRR458 pKa = 11.84KK459 pKa = 9.33YY460 pKa = 10.34GRR462 pKa = 11.84VSHH465 pKa = 6.5EE466 pKa = 4.1EE467 pKa = 3.85VPKK470 pKa = 9.45EE471 pKa = 3.62LRR473 pKa = 11.84VYY475 pKa = 10.48RR476 pKa = 11.84HH477 pKa = 5.74
MM1 pKa = 7.9ALRR4 pKa = 11.84SITGYY9 pKa = 10.15EE10 pKa = 3.73FHH12 pKa = 7.74DD13 pKa = 4.06FQSSLEE19 pKa = 4.3LLNQTHH25 pKa = 5.14IHH27 pKa = 5.36IVRR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 4.06SGVTYY37 pKa = 10.15HH38 pKa = 7.71DD39 pKa = 3.92EE40 pKa = 4.1FALCEE45 pKa = 4.03LLVDD49 pKa = 3.6NTRR52 pKa = 11.84LYY54 pKa = 10.15EE55 pKa = 3.97QEE57 pKa = 3.94LEE59 pKa = 4.18GWSRR63 pKa = 11.84SYY65 pKa = 10.01YY66 pKa = 10.1TGEE69 pKa = 3.75QHH71 pKa = 6.39MKK73 pKa = 10.8AILQYY78 pKa = 10.62SLPNTPIQDD87 pKa = 3.17IDD89 pKa = 3.78VGCYY93 pKa = 6.98QQAMTNVQEE102 pKa = 4.56RR103 pKa = 11.84LSSLPIVRR111 pKa = 11.84AFDD114 pKa = 3.43VLTQLDD120 pKa = 3.83QVSFEE125 pKa = 4.25SSSAAGYY132 pKa = 10.44DD133 pKa = 3.42YY134 pKa = 11.25TGAKK138 pKa = 9.43GPKK141 pKa = 9.39NEE143 pKa = 4.37GNHH146 pKa = 4.54EE147 pKa = 3.95RR148 pKa = 11.84AIRR151 pKa = 11.84RR152 pKa = 11.84AKK154 pKa = 9.99AVLWSAIAQDD164 pKa = 4.06GEE166 pKa = 4.64GIEE169 pKa = 4.26HH170 pKa = 6.53VLRR173 pKa = 11.84SSVPDD178 pKa = 3.17VGYY181 pKa = 8.77TRR183 pKa = 11.84TQLTDD188 pKa = 3.48LSEE191 pKa = 4.12KK192 pKa = 9.74TKK194 pKa = 10.6VRR196 pKa = 11.84GVWGRR201 pKa = 11.84AFHH204 pKa = 6.63YY205 pKa = 9.76ILPEE209 pKa = 4.15GTSADD214 pKa = 3.84PLLQAFKK221 pKa = 10.87EE222 pKa = 4.45GGTFYY227 pKa = 10.77HH228 pKa = 7.2IGQDD232 pKa = 3.49PTVSVPYY239 pKa = 10.01ILSDD243 pKa = 3.6TAGKK247 pKa = 9.67CAWLYY252 pKa = 11.14ALDD255 pKa = 3.85WSSFDD260 pKa = 3.26ATVSRR265 pKa = 11.84FEE267 pKa = 3.48IHH269 pKa = 7.07AAFDD273 pKa = 3.93LLKK276 pKa = 10.62QRR278 pKa = 11.84IEE280 pKa = 4.08FPNFEE285 pKa = 4.2TEE287 pKa = 3.56QCYY290 pKa = 9.72EE291 pKa = 3.47ICRR294 pKa = 11.84QLFIHH299 pKa = 6.58KK300 pKa = 9.99KK301 pKa = 8.91IAAPDD306 pKa = 3.68GKK308 pKa = 10.84VYY310 pKa = 9.56WAHH313 pKa = 6.77KK314 pKa = 8.99GIPSGSYY321 pKa = 6.92YY322 pKa = 10.46TSIIGSIINRR332 pKa = 11.84LRR334 pKa = 11.84IEE336 pKa = 4.37YY337 pKa = 9.63IWIKK341 pKa = 10.68LRR343 pKa = 11.84GHH345 pKa = 6.83GPTICYY351 pKa = 7.61TQGDD355 pKa = 4.29DD356 pKa = 4.56SLCGDD361 pKa = 4.47DD362 pKa = 5.6EE363 pKa = 5.59RR364 pKa = 11.84IEE366 pKa = 4.09PEE368 pKa = 4.58RR369 pKa = 11.84IADD372 pKa = 3.46IANPIGWLINPAKK385 pKa = 9.63TATTRR390 pKa = 11.84YY391 pKa = 9.02PEE393 pKa = 4.52YY394 pKa = 9.68ITFLGRR400 pKa = 11.84TCYY403 pKa = 10.48GGLNHH408 pKa = 7.54RR409 pKa = 11.84DD410 pKa = 3.64LIRR413 pKa = 11.84CLRR416 pKa = 11.84LLIYY420 pKa = 10.05PEE422 pKa = 4.19YY423 pKa = 9.12PVPSGAISAYY433 pKa = 8.96RR434 pKa = 11.84ANSIAEE440 pKa = 4.24DD441 pKa = 3.75CGGTSSILNDD451 pKa = 2.94IARR454 pKa = 11.84RR455 pKa = 11.84LTRR458 pKa = 11.84KK459 pKa = 9.33YY460 pKa = 10.34GRR462 pKa = 11.84VSHH465 pKa = 6.5EE466 pKa = 4.1EE467 pKa = 3.85VPKK470 pKa = 9.45EE471 pKa = 3.62LRR473 pKa = 11.84VYY475 pKa = 10.48RR476 pKa = 11.84HH477 pKa = 5.74
Molecular weight: 54.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4EPL9|I4EPL9_9VIRU Coat protein OS=Persimmon cryptic virus OX=1183241 GN=CP PE=4 SV=1
MM1 pKa = 7.94ADD3 pKa = 3.05QGNNEE8 pKa = 4.22TPLPVQATQPAANSDD23 pKa = 4.43RR24 pKa = 11.84IPITTLPAPRR34 pKa = 11.84SQAPNKK40 pKa = 9.78PEE42 pKa = 3.77YY43 pKa = 10.89ARR45 pKa = 11.84IADD48 pKa = 4.0SLEE51 pKa = 4.12DD52 pKa = 3.71EE53 pKa = 4.49YY54 pKa = 11.5QIGFQRR60 pKa = 11.84INPRR64 pKa = 11.84LHH66 pKa = 5.76RR67 pKa = 11.84RR68 pKa = 11.84YY69 pKa = 10.4VILRR73 pKa = 11.84SQEE76 pKa = 3.73LHH78 pKa = 7.43DD79 pKa = 4.51RR80 pKa = 11.84LCNLYY85 pKa = 10.58APYY88 pKa = 9.79FQAGYY93 pKa = 10.23RR94 pKa = 11.84VFKK97 pKa = 10.53HH98 pKa = 6.09VIEE101 pKa = 5.3AYY103 pKa = 9.25PLPAGYY109 pKa = 10.54ASQIAYY115 pKa = 7.69ATACYY120 pKa = 9.86VSSWCWDD127 pKa = 3.51LYY129 pKa = 10.04VSIRR133 pKa = 11.84EE134 pKa = 4.27SVKK137 pKa = 10.69KK138 pKa = 10.84LSQTAFLQQYY148 pKa = 6.6QTEE151 pKa = 4.4QFHH154 pKa = 6.07SQDD157 pKa = 3.19RR158 pKa = 11.84YY159 pKa = 11.57DD160 pKa = 4.73PFLQHH165 pKa = 6.96LNTVIRR171 pKa = 11.84PTHH174 pKa = 5.84IPGGLEE180 pKa = 3.76DD181 pKa = 4.13SLYY184 pKa = 10.52IPLIANHH191 pKa = 5.71VQWGNQSPFGINHH204 pKa = 5.36YY205 pKa = 10.26AINEE209 pKa = 4.24SLVQGLLDD217 pKa = 5.32VMDD220 pKa = 5.37LKK222 pKa = 11.49NNTWRR227 pKa = 11.84TVPLSHH233 pKa = 7.27DD234 pKa = 3.54TLGRR238 pKa = 11.84PMWLLDD244 pKa = 3.03WRR246 pKa = 11.84QNQAYY251 pKa = 9.98AWFPQEE257 pKa = 3.7NHH259 pKa = 6.09FSKK262 pKa = 10.93EE263 pKa = 3.84DD264 pKa = 3.83LIAAHH269 pKa = 7.09ILGIPCSPRR278 pKa = 11.84LGPRR282 pKa = 11.84DD283 pKa = 3.32VDD285 pKa = 3.13DD286 pKa = 3.75WQFFPGNVMPQHH298 pKa = 6.32INIAAYY304 pKa = 9.43EE305 pKa = 4.02RR306 pKa = 11.84AIPRR310 pKa = 11.84RR311 pKa = 11.84FFGAAEE317 pKa = 3.92YY318 pKa = 8.56RR319 pKa = 11.84TISTRR324 pKa = 11.84MWEE327 pKa = 4.44LPFLSAVAGQMSGAKK342 pKa = 9.71RR343 pKa = 11.84PAPEE347 pKa = 4.11SSATGSGYY355 pKa = 9.26MEE357 pKa = 4.74SPPAVEE363 pKa = 4.62GQQTAAIMDD372 pKa = 4.47PGIMQTRR379 pKa = 11.84YY380 pKa = 9.48CQYY383 pKa = 10.96QIIDD387 pKa = 3.29WCYY390 pKa = 8.43HH391 pKa = 4.39GRR393 pKa = 11.84VILNLNAQKK402 pKa = 10.58QNQALRR408 pKa = 11.84SILFSPNN415 pKa = 2.57
MM1 pKa = 7.94ADD3 pKa = 3.05QGNNEE8 pKa = 4.22TPLPVQATQPAANSDD23 pKa = 4.43RR24 pKa = 11.84IPITTLPAPRR34 pKa = 11.84SQAPNKK40 pKa = 9.78PEE42 pKa = 3.77YY43 pKa = 10.89ARR45 pKa = 11.84IADD48 pKa = 4.0SLEE51 pKa = 4.12DD52 pKa = 3.71EE53 pKa = 4.49YY54 pKa = 11.5QIGFQRR60 pKa = 11.84INPRR64 pKa = 11.84LHH66 pKa = 5.76RR67 pKa = 11.84RR68 pKa = 11.84YY69 pKa = 10.4VILRR73 pKa = 11.84SQEE76 pKa = 3.73LHH78 pKa = 7.43DD79 pKa = 4.51RR80 pKa = 11.84LCNLYY85 pKa = 10.58APYY88 pKa = 9.79FQAGYY93 pKa = 10.23RR94 pKa = 11.84VFKK97 pKa = 10.53HH98 pKa = 6.09VIEE101 pKa = 5.3AYY103 pKa = 9.25PLPAGYY109 pKa = 10.54ASQIAYY115 pKa = 7.69ATACYY120 pKa = 9.86VSSWCWDD127 pKa = 3.51LYY129 pKa = 10.04VSIRR133 pKa = 11.84EE134 pKa = 4.27SVKK137 pKa = 10.69KK138 pKa = 10.84LSQTAFLQQYY148 pKa = 6.6QTEE151 pKa = 4.4QFHH154 pKa = 6.07SQDD157 pKa = 3.19RR158 pKa = 11.84YY159 pKa = 11.57DD160 pKa = 4.73PFLQHH165 pKa = 6.96LNTVIRR171 pKa = 11.84PTHH174 pKa = 5.84IPGGLEE180 pKa = 3.76DD181 pKa = 4.13SLYY184 pKa = 10.52IPLIANHH191 pKa = 5.71VQWGNQSPFGINHH204 pKa = 5.36YY205 pKa = 10.26AINEE209 pKa = 4.24SLVQGLLDD217 pKa = 5.32VMDD220 pKa = 5.37LKK222 pKa = 11.49NNTWRR227 pKa = 11.84TVPLSHH233 pKa = 7.27DD234 pKa = 3.54TLGRR238 pKa = 11.84PMWLLDD244 pKa = 3.03WRR246 pKa = 11.84QNQAYY251 pKa = 9.98AWFPQEE257 pKa = 3.7NHH259 pKa = 6.09FSKK262 pKa = 10.93EE263 pKa = 3.84DD264 pKa = 3.83LIAAHH269 pKa = 7.09ILGIPCSPRR278 pKa = 11.84LGPRR282 pKa = 11.84DD283 pKa = 3.32VDD285 pKa = 3.13DD286 pKa = 3.75WQFFPGNVMPQHH298 pKa = 6.32INIAAYY304 pKa = 9.43EE305 pKa = 4.02RR306 pKa = 11.84AIPRR310 pKa = 11.84RR311 pKa = 11.84FFGAAEE317 pKa = 3.92YY318 pKa = 8.56RR319 pKa = 11.84TISTRR324 pKa = 11.84MWEE327 pKa = 4.44LPFLSAVAGQMSGAKK342 pKa = 9.71RR343 pKa = 11.84PAPEE347 pKa = 4.11SSATGSGYY355 pKa = 9.26MEE357 pKa = 4.74SPPAVEE363 pKa = 4.62GQQTAAIMDD372 pKa = 4.47PGIMQTRR379 pKa = 11.84YY380 pKa = 9.48CQYY383 pKa = 10.96QIIDD387 pKa = 3.29WCYY390 pKa = 8.43HH391 pKa = 4.39GRR393 pKa = 11.84VILNLNAQKK402 pKa = 10.58QNQALRR408 pKa = 11.84SILFSPNN415 pKa = 2.57
Molecular weight: 47.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
892 |
415 |
477 |
446.0 |
50.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.072 ± 0.697 | 1.794 ± 0.224 |
5.045 ± 0.3 | 5.717 ± 0.887 |
3.363 ± 0.161 | 6.278 ± 0.783 |
3.251 ± 0.076 | 7.511 ± 0.491 |
2.803 ± 0.562 | 8.408 ± 0.138 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.345 ± 0.529 | 3.812 ± 0.802 |
6.054 ± 1.22 | 5.83 ± 1.364 |
6.839 ± 0.214 | 6.726 ± 0.297 |
5.157 ± 0.682 | 4.372 ± 0.177 |
2.018 ± 0.252 | 5.605 ± 0.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
