
Bat Middle East Hepe-Astrovirus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
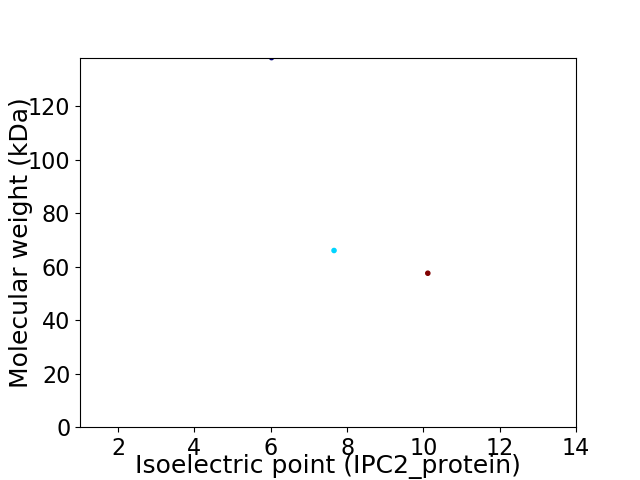
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
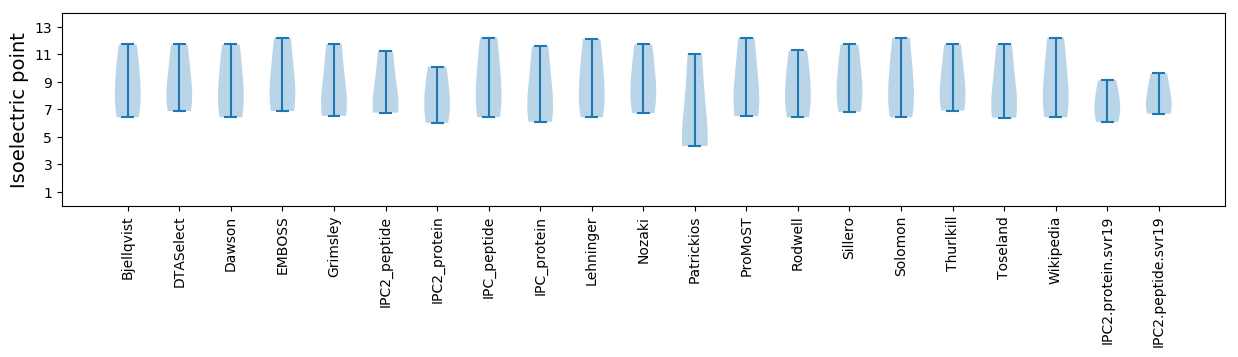
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U7N8C8|A0A2U7N8C8_9VIRU Capsid polyprotein VP90 OS=Bat Middle East Hepe-Astrovirus OX=2301732 PE=3 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84VLFFRR7 pKa = 11.84GSPPMSMTSSACRR20 pKa = 11.84SAMDD24 pKa = 4.14LSYY27 pKa = 10.68PQGAEE32 pKa = 4.27PTWGTQIAAINAGAAARR49 pKa = 11.84RR50 pKa = 11.84VRR52 pKa = 11.84AALHH56 pKa = 5.15VPYY59 pKa = 10.85VLTQTQKK66 pKa = 9.17TRR68 pKa = 11.84FKK70 pKa = 11.22DD71 pKa = 3.57LVRR74 pKa = 11.84CDD76 pKa = 3.45VVFEE80 pKa = 4.47DD81 pKa = 4.97RR82 pKa = 11.84VEE84 pKa = 4.09HH85 pKa = 4.89THH87 pKa = 6.03AVLRR91 pKa = 11.84ALHH94 pKa = 5.79YY95 pKa = 10.24FADD98 pKa = 3.79ATAFRR103 pKa = 11.84HH104 pKa = 5.97ADD106 pKa = 3.49DD107 pKa = 5.22VIDD110 pKa = 4.16IGGDD114 pKa = 3.14PRR116 pKa = 11.84KK117 pKa = 10.51NKK119 pKa = 7.64TRR121 pKa = 11.84RR122 pKa = 11.84NHH124 pKa = 4.0VCMMVDD130 pKa = 3.3NARR133 pKa = 11.84DD134 pKa = 3.35EE135 pKa = 4.31LRR137 pKa = 11.84AMRR140 pKa = 11.84ANLDD144 pKa = 3.39PRR146 pKa = 11.84PDD148 pKa = 3.26VGTRR152 pKa = 11.84CYY154 pKa = 10.92GGAQWCNVQAPRR166 pKa = 11.84GVSIHH171 pKa = 5.83SAYY174 pKa = 10.6DD175 pKa = 3.21ITWEE179 pKa = 3.7VWEE182 pKa = 4.84QIFEE186 pKa = 4.15AHH188 pKa = 6.98GLQCVDD194 pKa = 2.79VWLLEE199 pKa = 4.0PEE201 pKa = 4.22EE202 pKa = 4.56LFGGTTDD209 pKa = 3.25AFYY212 pKa = 10.81GIQTTAVGQNHH223 pKa = 5.87VMLLDD228 pKa = 4.95DD229 pKa = 3.95GTAGYY234 pKa = 7.52EE235 pKa = 3.94HH236 pKa = 7.06RR237 pKa = 11.84TVEE240 pKa = 3.75WTKK243 pKa = 9.5YY244 pKa = 9.27RR245 pKa = 11.84LSNGHH250 pKa = 5.44VGKK253 pKa = 10.23NFNLAVLPEE262 pKa = 4.35GQWGILRR269 pKa = 11.84RR270 pKa = 11.84YY271 pKa = 8.28TIFRR275 pKa = 11.84SHH277 pKa = 6.55RR278 pKa = 11.84PARR281 pKa = 11.84IWQAKK286 pKa = 8.65CAVDD290 pKa = 3.77RR291 pKa = 11.84NTAHH295 pKa = 6.51VPTLAGGCIKK305 pKa = 10.63VPLQHH310 pKa = 6.98WDD312 pKa = 3.52NLLSWGLARR321 pKa = 11.84GDD323 pKa = 3.62EE324 pKa = 4.32KK325 pKa = 11.03FTFQALMAYY334 pKa = 9.8ARR336 pKa = 11.84ALRR339 pKa = 11.84HH340 pKa = 5.4RR341 pKa = 11.84LIVGSNVVHH350 pKa = 6.9GGWTQDD356 pKa = 2.75AVEE359 pKa = 4.06QHH361 pKa = 7.01DD362 pKa = 4.28VVQTAFVVCAALRR375 pKa = 11.84FMRR378 pKa = 11.84TQSLSLALRR387 pKa = 11.84RR388 pKa = 11.84LKK390 pKa = 10.72HH391 pKa = 4.61EE392 pKa = 4.22QGRR395 pKa = 11.84GWFGEE400 pKa = 4.11LIDD403 pKa = 5.74RR404 pKa = 11.84LKK406 pKa = 11.08AWLGIPAIRR415 pKa = 11.84LRR417 pKa = 11.84QFLVTDD423 pKa = 3.56LHH425 pKa = 6.06NLRR428 pKa = 11.84AGRR431 pKa = 11.84DD432 pKa = 3.45SAVPLLRR439 pKa = 11.84EE440 pKa = 3.75LPGRR444 pKa = 11.84VGGNFWPEE452 pKa = 3.54EE453 pKa = 4.11DD454 pKa = 3.68TDD456 pKa = 3.87EE457 pKa = 4.94GPEE460 pKa = 4.0FTPEE464 pKa = 3.75EE465 pKa = 4.27PLPPEE470 pKa = 4.13PAEE473 pKa = 3.74LLAYY477 pKa = 8.06RR478 pKa = 11.84TALQAQTCTAGPYY491 pKa = 10.38AEE493 pKa = 5.5LAQQTLDD500 pKa = 3.61FLDD503 pKa = 3.99AAEE506 pKa = 4.34CRR508 pKa = 11.84ARR510 pKa = 11.84PKK512 pKa = 9.69RR513 pKa = 11.84TLIEE517 pKa = 4.37GPPGSGKK524 pKa = 7.47STRR527 pKa = 11.84FADD530 pKa = 3.24WSDD533 pKa = 3.29DD534 pKa = 3.4RR535 pKa = 11.84KK536 pKa = 11.2ANTLVVVPTKK546 pKa = 10.29KK547 pKa = 10.29QKK549 pKa = 11.06ADD551 pKa = 2.74WAQRR555 pKa = 11.84GFTAHH560 pKa = 6.23TPQRR564 pKa = 11.84ALQQCVGYY572 pKa = 9.09PLVIVDD578 pKa = 4.43EE579 pKa = 4.62VSLVHH584 pKa = 6.21PGVVRR589 pKa = 11.84ALEE592 pKa = 4.12RR593 pKa = 11.84HH594 pKa = 5.73SGAAQLYY601 pKa = 9.55CVGDD605 pKa = 3.83LKK607 pKa = 11.18QIAFCDD613 pKa = 4.25FEE615 pKa = 4.55GLGVEE620 pKa = 4.91FDD622 pKa = 3.78LQRR625 pKa = 11.84FYY627 pKa = 11.77ADD629 pKa = 3.39WEE631 pKa = 4.55KK632 pKa = 10.36EE633 pKa = 4.38TLTVTHH639 pKa = 7.3RR640 pKa = 11.84CPVDD644 pKa = 3.33VTKK647 pKa = 11.07VLGRR651 pKa = 11.84YY652 pKa = 9.0YY653 pKa = 10.7EE654 pKa = 5.12GITTTNTRR662 pKa = 11.84QVSIIKK668 pKa = 8.96GVCNPAAQHH677 pKa = 6.06LCYY680 pKa = 9.83TQGTKK685 pKa = 10.38SKK687 pKa = 9.4LTSQHH692 pKa = 6.17PNVLTVHH699 pKa = 6.53EE700 pKa = 4.64AQGGTYY706 pKa = 10.2KK707 pKa = 10.54AVCLHH712 pKa = 6.45VEE714 pKa = 4.12AQDD717 pKa = 3.35ATLVRR722 pKa = 11.84TSRR725 pKa = 11.84AHH727 pKa = 5.73NLVAITRR734 pKa = 11.84HH735 pKa = 5.05TGSLVVCEE743 pKa = 4.41QGTTALSEE751 pKa = 4.06WLDD754 pKa = 3.38PAFAVCEE761 pKa = 3.92LAEE764 pKa = 4.61GLDD767 pKa = 3.91AAPVTVPEE775 pKa = 4.4DD776 pKa = 3.21ARR778 pKa = 11.84ATVSPADD785 pKa = 3.64EE786 pKa = 5.19AGNSIPPHH794 pKa = 5.76TGANLDD800 pKa = 3.86DD801 pKa = 5.06LGVTFGADD809 pKa = 2.93LGVEE813 pKa = 4.76RR814 pKa = 11.84ITAAALPAAQRR825 pKa = 11.84TAQIQLDD832 pKa = 4.26SFPTLDD838 pKa = 3.38VSKK841 pKa = 10.61LALTAVPAPACRR853 pKa = 11.84FSRR856 pKa = 11.84PQDD859 pKa = 3.19KK860 pKa = 10.45AAAAAAVLSRR870 pKa = 11.84ITGTNKK876 pKa = 9.83NCGKK880 pKa = 10.2QEE882 pKa = 3.92AKK884 pKa = 10.62VRR886 pKa = 11.84GAALRR891 pKa = 11.84RR892 pKa = 11.84AFDD895 pKa = 3.15QWLEE899 pKa = 4.05PEE901 pKa = 4.55DD902 pKa = 4.67AVVSPDD908 pKa = 3.35EE909 pKa = 3.95QFLRR913 pKa = 11.84LAEE916 pKa = 4.2ALRR919 pKa = 11.84AAAEE923 pKa = 4.1KK924 pKa = 10.82DD925 pKa = 3.26PEE927 pKa = 4.23LRR929 pKa = 11.84RR930 pKa = 11.84LLAEE934 pKa = 4.46EE935 pKa = 4.7DD936 pKa = 3.65LVDD939 pKa = 4.61IIRR942 pKa = 11.84VEE944 pKa = 3.96NHH946 pKa = 6.55LKK948 pKa = 9.63QQAKK952 pKa = 9.65FAGEE956 pKa = 3.93PLRR959 pKa = 11.84KK960 pKa = 9.12IKK962 pKa = 10.58AGQGIDD968 pKa = 3.2AWSKK972 pKa = 10.27KK973 pKa = 9.79ANLVIGPFIRR983 pKa = 11.84AAQEE987 pKa = 3.57RR988 pKa = 11.84LLRR991 pKa = 11.84RR992 pKa = 11.84LKK994 pKa = 9.06PTVMMILHH1002 pKa = 6.04QSDD1005 pKa = 3.53ADD1007 pKa = 3.56VRR1009 pKa = 11.84DD1010 pKa = 4.44WIRR1013 pKa = 11.84SVHH1016 pKa = 5.67TGNTCVCNDD1025 pKa = 3.17FTEE1028 pKa = 4.7FDD1030 pKa = 3.65STQNDD1035 pKa = 3.38GTTRR1039 pKa = 11.84FEE1041 pKa = 4.44CLLLADD1047 pKa = 5.16LGVPKK1052 pKa = 10.32PIVNTYY1058 pKa = 9.44KK1059 pKa = 10.99ALRR1062 pKa = 11.84EE1063 pKa = 4.16SARR1066 pKa = 11.84VHH1068 pKa = 6.14ANGVVTDD1075 pKa = 4.04AAYY1078 pKa = 10.75ARR1080 pKa = 11.84MSGEE1084 pKa = 4.04ANTLFGNTVVTMAVNALLFPGAFDD1108 pKa = 3.35WAAFKK1113 pKa = 11.12GDD1115 pKa = 4.73DD1116 pKa = 5.22SIICNPRR1123 pKa = 11.84NRR1125 pKa = 11.84SDD1127 pKa = 3.66PDD1129 pKa = 3.83IISHH1133 pKa = 4.68QTGMQCKK1140 pKa = 9.44IEE1142 pKa = 4.04EE1143 pKa = 4.08QHH1145 pKa = 6.4VSEE1148 pKa = 5.08FVGLLISTDD1157 pKa = 3.33HH1158 pKa = 6.22VFPDD1162 pKa = 3.33LRR1164 pKa = 11.84RR1165 pKa = 11.84RR1166 pKa = 11.84VGKK1169 pKa = 8.71LTGRR1173 pKa = 11.84GYY1175 pKa = 9.98PGDD1178 pKa = 3.75PMARR1182 pKa = 11.84DD1183 pKa = 3.45QFSQSVRR1190 pKa = 11.84DD1191 pKa = 3.8TLALITPEE1199 pKa = 3.97STEE1202 pKa = 4.0ACILLNALAHH1212 pKa = 5.92HH1213 pKa = 6.66VSPAEE1218 pKa = 3.73AQVWYY1223 pKa = 9.82DD1224 pKa = 3.26VLIACVEE1231 pKa = 4.23GLGKK1235 pKa = 9.72PNRR1238 pKa = 11.84VEE1240 pKa = 4.7AIEE1243 pKa = 4.23ARR1245 pKa = 11.84LDD1247 pKa = 3.63EE1248 pKa = 5.03EE1249 pKa = 4.3II1250 pKa = 5.1
MM1 pKa = 7.58RR2 pKa = 11.84VLFFRR7 pKa = 11.84GSPPMSMTSSACRR20 pKa = 11.84SAMDD24 pKa = 4.14LSYY27 pKa = 10.68PQGAEE32 pKa = 4.27PTWGTQIAAINAGAAARR49 pKa = 11.84RR50 pKa = 11.84VRR52 pKa = 11.84AALHH56 pKa = 5.15VPYY59 pKa = 10.85VLTQTQKK66 pKa = 9.17TRR68 pKa = 11.84FKK70 pKa = 11.22DD71 pKa = 3.57LVRR74 pKa = 11.84CDD76 pKa = 3.45VVFEE80 pKa = 4.47DD81 pKa = 4.97RR82 pKa = 11.84VEE84 pKa = 4.09HH85 pKa = 4.89THH87 pKa = 6.03AVLRR91 pKa = 11.84ALHH94 pKa = 5.79YY95 pKa = 10.24FADD98 pKa = 3.79ATAFRR103 pKa = 11.84HH104 pKa = 5.97ADD106 pKa = 3.49DD107 pKa = 5.22VIDD110 pKa = 4.16IGGDD114 pKa = 3.14PRR116 pKa = 11.84KK117 pKa = 10.51NKK119 pKa = 7.64TRR121 pKa = 11.84RR122 pKa = 11.84NHH124 pKa = 4.0VCMMVDD130 pKa = 3.3NARR133 pKa = 11.84DD134 pKa = 3.35EE135 pKa = 4.31LRR137 pKa = 11.84AMRR140 pKa = 11.84ANLDD144 pKa = 3.39PRR146 pKa = 11.84PDD148 pKa = 3.26VGTRR152 pKa = 11.84CYY154 pKa = 10.92GGAQWCNVQAPRR166 pKa = 11.84GVSIHH171 pKa = 5.83SAYY174 pKa = 10.6DD175 pKa = 3.21ITWEE179 pKa = 3.7VWEE182 pKa = 4.84QIFEE186 pKa = 4.15AHH188 pKa = 6.98GLQCVDD194 pKa = 2.79VWLLEE199 pKa = 4.0PEE201 pKa = 4.22EE202 pKa = 4.56LFGGTTDD209 pKa = 3.25AFYY212 pKa = 10.81GIQTTAVGQNHH223 pKa = 5.87VMLLDD228 pKa = 4.95DD229 pKa = 3.95GTAGYY234 pKa = 7.52EE235 pKa = 3.94HH236 pKa = 7.06RR237 pKa = 11.84TVEE240 pKa = 3.75WTKK243 pKa = 9.5YY244 pKa = 9.27RR245 pKa = 11.84LSNGHH250 pKa = 5.44VGKK253 pKa = 10.23NFNLAVLPEE262 pKa = 4.35GQWGILRR269 pKa = 11.84RR270 pKa = 11.84YY271 pKa = 8.28TIFRR275 pKa = 11.84SHH277 pKa = 6.55RR278 pKa = 11.84PARR281 pKa = 11.84IWQAKK286 pKa = 8.65CAVDD290 pKa = 3.77RR291 pKa = 11.84NTAHH295 pKa = 6.51VPTLAGGCIKK305 pKa = 10.63VPLQHH310 pKa = 6.98WDD312 pKa = 3.52NLLSWGLARR321 pKa = 11.84GDD323 pKa = 3.62EE324 pKa = 4.32KK325 pKa = 11.03FTFQALMAYY334 pKa = 9.8ARR336 pKa = 11.84ALRR339 pKa = 11.84HH340 pKa = 5.4RR341 pKa = 11.84LIVGSNVVHH350 pKa = 6.9GGWTQDD356 pKa = 2.75AVEE359 pKa = 4.06QHH361 pKa = 7.01DD362 pKa = 4.28VVQTAFVVCAALRR375 pKa = 11.84FMRR378 pKa = 11.84TQSLSLALRR387 pKa = 11.84RR388 pKa = 11.84LKK390 pKa = 10.72HH391 pKa = 4.61EE392 pKa = 4.22QGRR395 pKa = 11.84GWFGEE400 pKa = 4.11LIDD403 pKa = 5.74RR404 pKa = 11.84LKK406 pKa = 11.08AWLGIPAIRR415 pKa = 11.84LRR417 pKa = 11.84QFLVTDD423 pKa = 3.56LHH425 pKa = 6.06NLRR428 pKa = 11.84AGRR431 pKa = 11.84DD432 pKa = 3.45SAVPLLRR439 pKa = 11.84EE440 pKa = 3.75LPGRR444 pKa = 11.84VGGNFWPEE452 pKa = 3.54EE453 pKa = 4.11DD454 pKa = 3.68TDD456 pKa = 3.87EE457 pKa = 4.94GPEE460 pKa = 4.0FTPEE464 pKa = 3.75EE465 pKa = 4.27PLPPEE470 pKa = 4.13PAEE473 pKa = 3.74LLAYY477 pKa = 8.06RR478 pKa = 11.84TALQAQTCTAGPYY491 pKa = 10.38AEE493 pKa = 5.5LAQQTLDD500 pKa = 3.61FLDD503 pKa = 3.99AAEE506 pKa = 4.34CRR508 pKa = 11.84ARR510 pKa = 11.84PKK512 pKa = 9.69RR513 pKa = 11.84TLIEE517 pKa = 4.37GPPGSGKK524 pKa = 7.47STRR527 pKa = 11.84FADD530 pKa = 3.24WSDD533 pKa = 3.29DD534 pKa = 3.4RR535 pKa = 11.84KK536 pKa = 11.2ANTLVVVPTKK546 pKa = 10.29KK547 pKa = 10.29QKK549 pKa = 11.06ADD551 pKa = 2.74WAQRR555 pKa = 11.84GFTAHH560 pKa = 6.23TPQRR564 pKa = 11.84ALQQCVGYY572 pKa = 9.09PLVIVDD578 pKa = 4.43EE579 pKa = 4.62VSLVHH584 pKa = 6.21PGVVRR589 pKa = 11.84ALEE592 pKa = 4.12RR593 pKa = 11.84HH594 pKa = 5.73SGAAQLYY601 pKa = 9.55CVGDD605 pKa = 3.83LKK607 pKa = 11.18QIAFCDD613 pKa = 4.25FEE615 pKa = 4.55GLGVEE620 pKa = 4.91FDD622 pKa = 3.78LQRR625 pKa = 11.84FYY627 pKa = 11.77ADD629 pKa = 3.39WEE631 pKa = 4.55KK632 pKa = 10.36EE633 pKa = 4.38TLTVTHH639 pKa = 7.3RR640 pKa = 11.84CPVDD644 pKa = 3.33VTKK647 pKa = 11.07VLGRR651 pKa = 11.84YY652 pKa = 9.0YY653 pKa = 10.7EE654 pKa = 5.12GITTTNTRR662 pKa = 11.84QVSIIKK668 pKa = 8.96GVCNPAAQHH677 pKa = 6.06LCYY680 pKa = 9.83TQGTKK685 pKa = 10.38SKK687 pKa = 9.4LTSQHH692 pKa = 6.17PNVLTVHH699 pKa = 6.53EE700 pKa = 4.64AQGGTYY706 pKa = 10.2KK707 pKa = 10.54AVCLHH712 pKa = 6.45VEE714 pKa = 4.12AQDD717 pKa = 3.35ATLVRR722 pKa = 11.84TSRR725 pKa = 11.84AHH727 pKa = 5.73NLVAITRR734 pKa = 11.84HH735 pKa = 5.05TGSLVVCEE743 pKa = 4.41QGTTALSEE751 pKa = 4.06WLDD754 pKa = 3.38PAFAVCEE761 pKa = 3.92LAEE764 pKa = 4.61GLDD767 pKa = 3.91AAPVTVPEE775 pKa = 4.4DD776 pKa = 3.21ARR778 pKa = 11.84ATVSPADD785 pKa = 3.64EE786 pKa = 5.19AGNSIPPHH794 pKa = 5.76TGANLDD800 pKa = 3.86DD801 pKa = 5.06LGVTFGADD809 pKa = 2.93LGVEE813 pKa = 4.76RR814 pKa = 11.84ITAAALPAAQRR825 pKa = 11.84TAQIQLDD832 pKa = 4.26SFPTLDD838 pKa = 3.38VSKK841 pKa = 10.61LALTAVPAPACRR853 pKa = 11.84FSRR856 pKa = 11.84PQDD859 pKa = 3.19KK860 pKa = 10.45AAAAAAVLSRR870 pKa = 11.84ITGTNKK876 pKa = 9.83NCGKK880 pKa = 10.2QEE882 pKa = 3.92AKK884 pKa = 10.62VRR886 pKa = 11.84GAALRR891 pKa = 11.84RR892 pKa = 11.84AFDD895 pKa = 3.15QWLEE899 pKa = 4.05PEE901 pKa = 4.55DD902 pKa = 4.67AVVSPDD908 pKa = 3.35EE909 pKa = 3.95QFLRR913 pKa = 11.84LAEE916 pKa = 4.2ALRR919 pKa = 11.84AAAEE923 pKa = 4.1KK924 pKa = 10.82DD925 pKa = 3.26PEE927 pKa = 4.23LRR929 pKa = 11.84RR930 pKa = 11.84LLAEE934 pKa = 4.46EE935 pKa = 4.7DD936 pKa = 3.65LVDD939 pKa = 4.61IIRR942 pKa = 11.84VEE944 pKa = 3.96NHH946 pKa = 6.55LKK948 pKa = 9.63QQAKK952 pKa = 9.65FAGEE956 pKa = 3.93PLRR959 pKa = 11.84KK960 pKa = 9.12IKK962 pKa = 10.58AGQGIDD968 pKa = 3.2AWSKK972 pKa = 10.27KK973 pKa = 9.79ANLVIGPFIRR983 pKa = 11.84AAQEE987 pKa = 3.57RR988 pKa = 11.84LLRR991 pKa = 11.84RR992 pKa = 11.84LKK994 pKa = 9.06PTVMMILHH1002 pKa = 6.04QSDD1005 pKa = 3.53ADD1007 pKa = 3.56VRR1009 pKa = 11.84DD1010 pKa = 4.44WIRR1013 pKa = 11.84SVHH1016 pKa = 5.67TGNTCVCNDD1025 pKa = 3.17FTEE1028 pKa = 4.7FDD1030 pKa = 3.65STQNDD1035 pKa = 3.38GTTRR1039 pKa = 11.84FEE1041 pKa = 4.44CLLLADD1047 pKa = 5.16LGVPKK1052 pKa = 10.32PIVNTYY1058 pKa = 9.44KK1059 pKa = 10.99ALRR1062 pKa = 11.84EE1063 pKa = 4.16SARR1066 pKa = 11.84VHH1068 pKa = 6.14ANGVVTDD1075 pKa = 4.04AAYY1078 pKa = 10.75ARR1080 pKa = 11.84MSGEE1084 pKa = 4.04ANTLFGNTVVTMAVNALLFPGAFDD1108 pKa = 3.35WAAFKK1113 pKa = 11.12GDD1115 pKa = 4.73DD1116 pKa = 5.22SIICNPRR1123 pKa = 11.84NRR1125 pKa = 11.84SDD1127 pKa = 3.66PDD1129 pKa = 3.83IISHH1133 pKa = 4.68QTGMQCKK1140 pKa = 9.44IEE1142 pKa = 4.04EE1143 pKa = 4.08QHH1145 pKa = 6.4VSEE1148 pKa = 5.08FVGLLISTDD1157 pKa = 3.33HH1158 pKa = 6.22VFPDD1162 pKa = 3.33LRR1164 pKa = 11.84RR1165 pKa = 11.84RR1166 pKa = 11.84VGKK1169 pKa = 8.71LTGRR1173 pKa = 11.84GYY1175 pKa = 9.98PGDD1178 pKa = 3.75PMARR1182 pKa = 11.84DD1183 pKa = 3.45QFSQSVRR1190 pKa = 11.84DD1191 pKa = 3.8TLALITPEE1199 pKa = 3.97STEE1202 pKa = 4.0ACILLNALAHH1212 pKa = 5.92HH1213 pKa = 6.66VSPAEE1218 pKa = 3.73AQVWYY1223 pKa = 9.82DD1224 pKa = 3.26VLIACVEE1231 pKa = 4.23GLGKK1235 pKa = 9.72PNRR1238 pKa = 11.84VEE1240 pKa = 4.7AIEE1243 pKa = 4.23ARR1245 pKa = 11.84LDD1247 pKa = 3.63EE1248 pKa = 5.03EE1249 pKa = 4.3II1250 pKa = 5.1
Molecular weight: 138.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U7MWY3|A0A2U7MWY3_9VIRU ORF1 OS=Bat Middle East Hepe-Astrovirus OX=2301732 PE=4 SV=1
MM1 pKa = 7.91PFRR4 pKa = 11.84MQGSRR9 pKa = 11.84SSSGRR14 pKa = 11.84ARR16 pKa = 11.84DD17 pKa = 3.46QRR19 pKa = 11.84SRR21 pKa = 11.84NLGQPRR27 pKa = 11.84LISARR32 pKa = 11.84SLARR36 pKa = 11.84TQMASPGSLTCSCTLLFCGTSMLRR60 pKa = 11.84TPPTPRR66 pKa = 11.84RR67 pKa = 11.84SEE69 pKa = 3.96RR70 pKa = 11.84RR71 pKa = 11.84STPCTVSQTWSLPCTAWWAPLASLVPSVSLRR102 pKa = 11.84CXLTLPRR109 pKa = 11.84AQPFPSTPSPRR120 pKa = 11.84ASTRR124 pKa = 11.84RR125 pKa = 11.84SASDD129 pKa = 3.11EE130 pKa = 4.13TDD132 pKa = 3.12GSNLAFHH139 pKa = 7.23PVRR142 pKa = 11.84ILGSSPTPRR151 pKa = 11.84RR152 pKa = 11.84TRR154 pKa = 11.84GPSRR158 pKa = 11.84SGKK161 pKa = 8.85RR162 pKa = 11.84WRR164 pKa = 11.84CSCLGQQTISTPTSPTLVHH183 pKa = 7.05FGDD186 pKa = 4.2SRR188 pKa = 11.84CGCVTSSPTTSRR200 pKa = 11.84TSSWRR205 pKa = 11.84LLRR208 pKa = 11.84QGRR211 pKa = 11.84RR212 pKa = 11.84LILPRR217 pKa = 11.84WPRR220 pKa = 11.84MRR222 pKa = 11.84PGTSQSRR229 pKa = 11.84QQSLSSAHH237 pKa = 6.62GSTHH241 pKa = 6.84PPQEE245 pKa = 4.44LPMSCSPSLTSQVGSLLKK263 pKa = 10.27FQSSAHH269 pKa = 6.64CSTQGWLSSSRR280 pKa = 11.84CSARR284 pKa = 11.84PGTGRR289 pKa = 11.84TSTSCAPVSTRR300 pKa = 11.84PRR302 pKa = 11.84LATPSQFQMPSTPQSPEE319 pKa = 3.05ISACNSSPPPTSVAPPKK336 pKa = 10.18PPLPRR341 pKa = 11.84RR342 pKa = 11.84PPAPSSCPPPPSAWLRR358 pKa = 11.84GQPATSRR365 pKa = 11.84SPSPRR370 pKa = 11.84GRR372 pKa = 11.84ARR374 pKa = 11.84WNEE377 pKa = 4.03LVRR380 pKa = 11.84QATTASLSLTPSPAAPPSYY399 pKa = 10.66HH400 pKa = 6.98PVTPPISPYY409 pKa = 10.64AVSGPCGATEE419 pKa = 4.1TRR421 pKa = 11.84PSSSGSYY428 pKa = 10.52VPMGRR433 pKa = 11.84PAEE436 pKa = 4.11RR437 pKa = 11.84TAPSSTSPPTRR448 pKa = 11.84RR449 pKa = 11.84GPWVSSVRR457 pKa = 11.84QQLLLTSPGYY467 pKa = 11.08CPMCHH472 pKa = 6.74RR473 pKa = 11.84ATIAWSLGGQRR484 pKa = 11.84SPPPRR489 pKa = 11.84VCPRR493 pKa = 11.84MYY495 pKa = 11.08DD496 pKa = 3.41PVTRR500 pKa = 11.84SYY502 pKa = 11.48QCISTTGSHH511 pKa = 5.96HH512 pKa = 5.15QTRR515 pKa = 11.84QNGRR519 pKa = 11.84PRR521 pKa = 11.84TAGCFADD528 pKa = 4.14LQRR531 pKa = 11.84SVSLL535 pKa = 3.84
MM1 pKa = 7.91PFRR4 pKa = 11.84MQGSRR9 pKa = 11.84SSSGRR14 pKa = 11.84ARR16 pKa = 11.84DD17 pKa = 3.46QRR19 pKa = 11.84SRR21 pKa = 11.84NLGQPRR27 pKa = 11.84LISARR32 pKa = 11.84SLARR36 pKa = 11.84TQMASPGSLTCSCTLLFCGTSMLRR60 pKa = 11.84TPPTPRR66 pKa = 11.84RR67 pKa = 11.84SEE69 pKa = 3.96RR70 pKa = 11.84RR71 pKa = 11.84STPCTVSQTWSLPCTAWWAPLASLVPSVSLRR102 pKa = 11.84CXLTLPRR109 pKa = 11.84AQPFPSTPSPRR120 pKa = 11.84ASTRR124 pKa = 11.84RR125 pKa = 11.84SASDD129 pKa = 3.11EE130 pKa = 4.13TDD132 pKa = 3.12GSNLAFHH139 pKa = 7.23PVRR142 pKa = 11.84ILGSSPTPRR151 pKa = 11.84RR152 pKa = 11.84TRR154 pKa = 11.84GPSRR158 pKa = 11.84SGKK161 pKa = 8.85RR162 pKa = 11.84WRR164 pKa = 11.84CSCLGQQTISTPTSPTLVHH183 pKa = 7.05FGDD186 pKa = 4.2SRR188 pKa = 11.84CGCVTSSPTTSRR200 pKa = 11.84TSSWRR205 pKa = 11.84LLRR208 pKa = 11.84QGRR211 pKa = 11.84RR212 pKa = 11.84LILPRR217 pKa = 11.84WPRR220 pKa = 11.84MRR222 pKa = 11.84PGTSQSRR229 pKa = 11.84QQSLSSAHH237 pKa = 6.62GSTHH241 pKa = 6.84PPQEE245 pKa = 4.44LPMSCSPSLTSQVGSLLKK263 pKa = 10.27FQSSAHH269 pKa = 6.64CSTQGWLSSSRR280 pKa = 11.84CSARR284 pKa = 11.84PGTGRR289 pKa = 11.84TSTSCAPVSTRR300 pKa = 11.84PRR302 pKa = 11.84LATPSQFQMPSTPQSPEE319 pKa = 3.05ISACNSSPPPTSVAPPKK336 pKa = 10.18PPLPRR341 pKa = 11.84RR342 pKa = 11.84PPAPSSCPPPPSAWLRR358 pKa = 11.84GQPATSRR365 pKa = 11.84SPSPRR370 pKa = 11.84GRR372 pKa = 11.84ARR374 pKa = 11.84WNEE377 pKa = 4.03LVRR380 pKa = 11.84QATTASLSLTPSPAAPPSYY399 pKa = 10.66HH400 pKa = 6.98PVTPPISPYY409 pKa = 10.64AVSGPCGATEE419 pKa = 4.1TRR421 pKa = 11.84PSSSGSYY428 pKa = 10.52VPMGRR433 pKa = 11.84PAEE436 pKa = 4.11RR437 pKa = 11.84TAPSSTSPPTRR448 pKa = 11.84RR449 pKa = 11.84GPWVSSVRR457 pKa = 11.84QQLLLTSPGYY467 pKa = 11.08CPMCHH472 pKa = 6.74RR473 pKa = 11.84ATIAWSLGGQRR484 pKa = 11.84SPPPRR489 pKa = 11.84VCPRR493 pKa = 11.84MYY495 pKa = 11.08DD496 pKa = 3.41PVTRR500 pKa = 11.84SYY502 pKa = 11.48QCISTTGSHH511 pKa = 5.96HH512 pKa = 5.15QTRR515 pKa = 11.84QNGRR519 pKa = 11.84PRR521 pKa = 11.84TAGCFADD528 pKa = 4.14LQRR531 pKa = 11.84SVSLL535 pKa = 3.84
Molecular weight: 57.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2403 |
535 |
1250 |
801.0 |
87.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.154 ± 1.29 | 2.289 ± 0.733 |
4.786 ± 1.241 | 4.12 ± 1.107 |
3.121 ± 0.539 | 7.158 ± 0.649 |
2.538 ± 0.374 | 3.204 ± 0.565 |
2.871 ± 0.766 | 8.656 ± 0.504 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.415 ± 0.15 | 2.58 ± 0.543 |
7.574 ± 2.338 | 4.702 ± 0.188 |
7.948 ± 1.484 | 7.824 ± 3.322 |
8.656 ± 1.196 | 6.617 ± 1.119 |
1.914 ± 0.053 | 1.789 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
