
Streptococcus gordonii (strain Challis / ATCC 35105 / BCRC 15272 / CH1 / DL1 / V288)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus gordonii; Streptococcus gordonii str. Challis
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
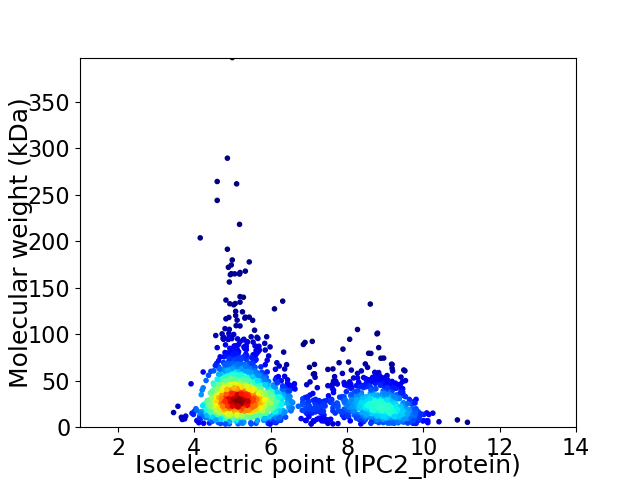
Virtual 2D-PAGE plot for 2050 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8AWL4|A8AWL4_STRGC ABC transporter ATP-binding protein OS=Streptococcus gordonii (strain Challis / ATCC 35105 / BCRC 15272 / CH1 / DL1 / V288) OX=467705 GN=SGO_0878 PE=4 SV=1
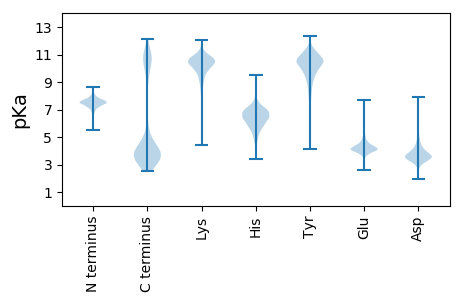
MM1 pKa = 7.54LSAIEE6 pKa = 4.3AQDD9 pKa = 4.05LVQAEE14 pKa = 4.18QWFEE18 pKa = 3.8KK19 pKa = 10.93ALLEE23 pKa = 4.89DD24 pKa = 4.02SDD26 pKa = 6.15SILLDD31 pKa = 3.36LAEE34 pKa = 4.27YY35 pKa = 10.55LEE37 pKa = 4.9SIGFFLQAKK46 pKa = 9.75RR47 pKa = 11.84IYY49 pKa = 10.68NKK51 pKa = 10.12LAPLYY56 pKa = 9.5PQVNLNLAAIASEE69 pKa = 4.67DD70 pKa = 3.66GDD72 pKa = 3.91LEE74 pKa = 4.2EE75 pKa = 4.84AFGYY79 pKa = 10.63LEE81 pKa = 4.98EE82 pKa = 5.09IRR84 pKa = 11.84SNDD87 pKa = 2.84PWYY90 pKa = 11.48VNALLIKK97 pKa = 10.25ADD99 pKa = 4.48LYY101 pKa = 11.42QMEE104 pKa = 4.82GLTDD108 pKa = 3.38VARR111 pKa = 11.84EE112 pKa = 3.98KK113 pKa = 10.85LVEE116 pKa = 3.98ASQLSDD122 pKa = 3.88DD123 pKa = 4.87PIITFGMAEE132 pKa = 3.96IDD134 pKa = 3.8FEE136 pKa = 5.22LGNFTQAIKK145 pKa = 10.43EE146 pKa = 4.12YY147 pKa = 10.81ASLDD151 pKa = 3.12NRR153 pKa = 11.84TIYY156 pKa = 10.09EE157 pKa = 3.78QTGISTYY164 pKa = 8.97QRR166 pKa = 11.84IGVSYY171 pKa = 10.58ASLGKK176 pKa = 10.23FEE178 pKa = 4.47AAIEE182 pKa = 3.97FLQKK186 pKa = 10.61AIEE189 pKa = 4.07LEE191 pKa = 4.09YY192 pKa = 11.23DD193 pKa = 3.24EE194 pKa = 5.59SAVFEE199 pKa = 4.48LAVILYY205 pKa = 9.62DD206 pKa = 4.08QEE208 pKa = 5.67DD209 pKa = 4.02YY210 pKa = 11.49QKK212 pKa = 11.61ANLYY216 pKa = 9.62FKK218 pKa = 10.66QLDD221 pKa = 3.78TLSPDD226 pKa = 3.33FEE228 pKa = 5.45GYY230 pKa = 10.42EE231 pKa = 4.02YY232 pKa = 10.09IYY234 pKa = 11.05ALSLHH239 pKa = 7.05ADD241 pKa = 3.63NQAEE245 pKa = 4.1AALLIAQQGLNKK257 pKa = 10.36NPFDD261 pKa = 3.81SQLALLASQVSYY273 pKa = 10.66EE274 pKa = 3.98LHH276 pKa = 6.08DD277 pKa = 3.74TEE279 pKa = 5.15KK280 pKa = 11.23AEE282 pKa = 5.3SYY284 pKa = 11.31LLDD287 pKa = 3.57AWDD290 pKa = 4.04NADD293 pKa = 4.81DD294 pKa = 4.7LEE296 pKa = 4.24EE297 pKa = 4.05LALRR301 pKa = 11.84LTNLYY306 pKa = 10.37LEE308 pKa = 4.28QEE310 pKa = 4.23RR311 pKa = 11.84YY312 pKa = 9.88EE313 pKa = 5.19DD314 pKa = 3.7ILALEE319 pKa = 4.59DD320 pKa = 3.87QDD322 pKa = 3.26WDD324 pKa = 3.77NVLTRR329 pKa = 11.84WNIARR334 pKa = 11.84SYY336 pKa = 10.33QALEE340 pKa = 4.94DD341 pKa = 3.59IEE343 pKa = 4.64KK344 pKa = 10.56AQEE347 pKa = 4.09LYY349 pKa = 11.41GEE351 pKa = 4.28LHH353 pKa = 6.96RR354 pKa = 11.84DD355 pKa = 3.25LRR357 pKa = 11.84DD358 pKa = 3.34NPEE361 pKa = 3.68FLEE364 pKa = 4.32EE365 pKa = 4.04YY366 pKa = 10.05IYY368 pKa = 10.97LLRR371 pKa = 11.84EE372 pKa = 4.19CGEE375 pKa = 4.31LEE377 pKa = 4.22SAKK380 pKa = 10.47QEE382 pKa = 4.08AQHH385 pKa = 5.92YY386 pKa = 9.59LSLVPDD392 pKa = 4.65DD393 pKa = 4.68SAMQEE398 pKa = 4.58FYY400 pKa = 11.35DD401 pKa = 4.11SLDD404 pKa = 3.53YY405 pKa = 11.4
MM1 pKa = 7.54LSAIEE6 pKa = 4.3AQDD9 pKa = 4.05LVQAEE14 pKa = 4.18QWFEE18 pKa = 3.8KK19 pKa = 10.93ALLEE23 pKa = 4.89DD24 pKa = 4.02SDD26 pKa = 6.15SILLDD31 pKa = 3.36LAEE34 pKa = 4.27YY35 pKa = 10.55LEE37 pKa = 4.9SIGFFLQAKK46 pKa = 9.75RR47 pKa = 11.84IYY49 pKa = 10.68NKK51 pKa = 10.12LAPLYY56 pKa = 9.5PQVNLNLAAIASEE69 pKa = 4.67DD70 pKa = 3.66GDD72 pKa = 3.91LEE74 pKa = 4.2EE75 pKa = 4.84AFGYY79 pKa = 10.63LEE81 pKa = 4.98EE82 pKa = 5.09IRR84 pKa = 11.84SNDD87 pKa = 2.84PWYY90 pKa = 11.48VNALLIKK97 pKa = 10.25ADD99 pKa = 4.48LYY101 pKa = 11.42QMEE104 pKa = 4.82GLTDD108 pKa = 3.38VARR111 pKa = 11.84EE112 pKa = 3.98KK113 pKa = 10.85LVEE116 pKa = 3.98ASQLSDD122 pKa = 3.88DD123 pKa = 4.87PIITFGMAEE132 pKa = 3.96IDD134 pKa = 3.8FEE136 pKa = 5.22LGNFTQAIKK145 pKa = 10.43EE146 pKa = 4.12YY147 pKa = 10.81ASLDD151 pKa = 3.12NRR153 pKa = 11.84TIYY156 pKa = 10.09EE157 pKa = 3.78QTGISTYY164 pKa = 8.97QRR166 pKa = 11.84IGVSYY171 pKa = 10.58ASLGKK176 pKa = 10.23FEE178 pKa = 4.47AAIEE182 pKa = 3.97FLQKK186 pKa = 10.61AIEE189 pKa = 4.07LEE191 pKa = 4.09YY192 pKa = 11.23DD193 pKa = 3.24EE194 pKa = 5.59SAVFEE199 pKa = 4.48LAVILYY205 pKa = 9.62DD206 pKa = 4.08QEE208 pKa = 5.67DD209 pKa = 4.02YY210 pKa = 11.49QKK212 pKa = 11.61ANLYY216 pKa = 9.62FKK218 pKa = 10.66QLDD221 pKa = 3.78TLSPDD226 pKa = 3.33FEE228 pKa = 5.45GYY230 pKa = 10.42EE231 pKa = 4.02YY232 pKa = 10.09IYY234 pKa = 11.05ALSLHH239 pKa = 7.05ADD241 pKa = 3.63NQAEE245 pKa = 4.1AALLIAQQGLNKK257 pKa = 10.36NPFDD261 pKa = 3.81SQLALLASQVSYY273 pKa = 10.66EE274 pKa = 3.98LHH276 pKa = 6.08DD277 pKa = 3.74TEE279 pKa = 5.15KK280 pKa = 11.23AEE282 pKa = 5.3SYY284 pKa = 11.31LLDD287 pKa = 3.57AWDD290 pKa = 4.04NADD293 pKa = 4.81DD294 pKa = 4.7LEE296 pKa = 4.24EE297 pKa = 4.05LALRR301 pKa = 11.84LTNLYY306 pKa = 10.37LEE308 pKa = 4.28QEE310 pKa = 4.23RR311 pKa = 11.84YY312 pKa = 9.88EE313 pKa = 5.19DD314 pKa = 3.7ILALEE319 pKa = 4.59DD320 pKa = 3.87QDD322 pKa = 3.26WDD324 pKa = 3.77NVLTRR329 pKa = 11.84WNIARR334 pKa = 11.84SYY336 pKa = 10.33QALEE340 pKa = 4.94DD341 pKa = 3.59IEE343 pKa = 4.64KK344 pKa = 10.56AQEE347 pKa = 4.09LYY349 pKa = 11.41GEE351 pKa = 4.28LHH353 pKa = 6.96RR354 pKa = 11.84DD355 pKa = 3.25LRR357 pKa = 11.84DD358 pKa = 3.34NPEE361 pKa = 3.68FLEE364 pKa = 4.32EE365 pKa = 4.04YY366 pKa = 10.05IYY368 pKa = 10.97LLRR371 pKa = 11.84EE372 pKa = 4.19CGEE375 pKa = 4.31LEE377 pKa = 4.22SAKK380 pKa = 10.47QEE382 pKa = 4.08AQHH385 pKa = 5.92YY386 pKa = 9.59LSLVPDD392 pKa = 4.65DD393 pKa = 4.68SAMQEE398 pKa = 4.58FYY400 pKa = 11.35DD401 pKa = 4.11SLDD404 pKa = 3.53YY405 pKa = 11.4
Molecular weight: 46.65 kDa
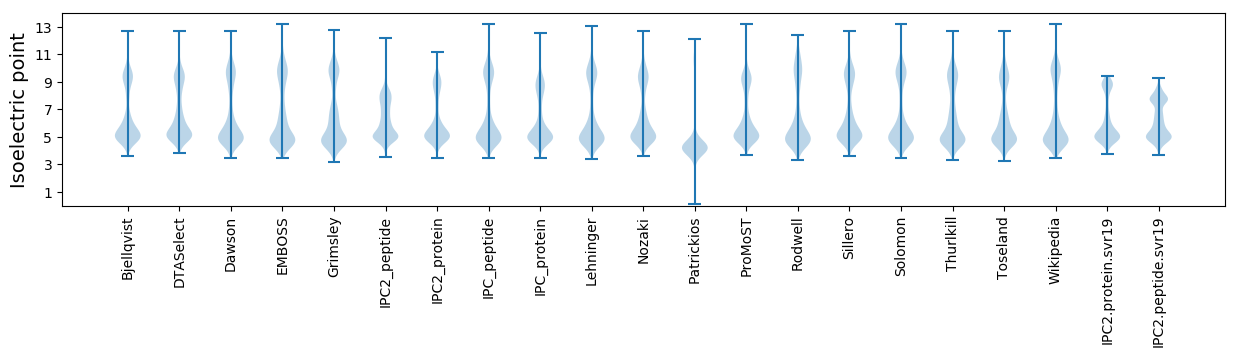
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8AUQ7|A8AUQ7_STRGC Small ribosomal subunit biogenesis GTPase RsgA OS=Streptococcus gordonii (strain Challis / ATCC 35105 / BCRC 15272 / CH1 / DL1 / V288) OX=467705 GN=rsgA PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.18IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.35HH16 pKa = 4.78GFRR19 pKa = 11.84NRR21 pKa = 11.84MSTKK25 pKa = 9.22NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.87GRR39 pKa = 11.84KK40 pKa = 8.75VLAAA44 pKa = 4.31
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.18IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.35HH16 pKa = 4.78GFRR19 pKa = 11.84NRR21 pKa = 11.84MSTKK25 pKa = 9.22NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.87GRR39 pKa = 11.84KK40 pKa = 8.75VLAAA44 pKa = 4.31
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
642221 |
30 |
3646 |
313.3 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.668 ± 0.067 | 0.526 ± 0.016 |
5.646 ± 0.049 | 7.196 ± 0.061 |
4.62 ± 0.054 | 6.564 ± 0.056 |
1.83 ± 0.022 | 7.067 ± 0.074 |
6.868 ± 0.061 | 10.223 ± 0.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.029 | 4.356 ± 0.043 |
3.424 ± 0.052 | 4.202 ± 0.046 |
3.968 ± 0.041 | 6.377 ± 0.109 |
5.533 ± 0.081 | 6.875 ± 0.049 |
0.905 ± 0.018 | 3.797 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
