
Papaya leaf curl alphasatellite
Taxonomy: Viruses; Alphasatellitidae; Geminialphasatellitinae; unclassified Begomovirus-associated alphasatellites
Average proteome isoelectric point is 8.82
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V9I5Y2|V9I5Y2_9VIRU Rep protein OS=Papaya leaf curl alphasatellite OX=1112202 PE=4 SV=1
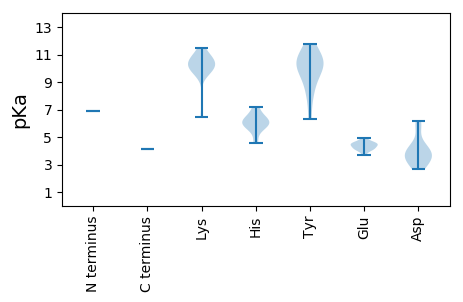
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTSLPEE24 pKa = 3.69WANYY28 pKa = 10.56LIFQEE33 pKa = 4.94EE34 pKa = 4.56EE35 pKa = 4.2CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.45VNLNAINDD55 pKa = 3.67LRR57 pKa = 11.84SCKK60 pKa = 10.1RR61 pKa = 11.84NCQMGLISKK70 pKa = 9.63HH71 pKa = 6.19AGVHH75 pKa = 5.71HH76 pKa = 6.53PAIEE80 pKa = 4.6IIAEE84 pKa = 4.07RR85 pKa = 11.84MIQEE89 pKa = 4.37STVHH93 pKa = 6.17GSSEE97 pKa = 4.18SLRR100 pKa = 11.84KK101 pKa = 9.84QGATRR106 pKa = 11.84GRR108 pKa = 11.84RR109 pKa = 11.84WNVFRR114 pKa = 11.84KK115 pKa = 6.43TQKK118 pKa = 10.89NLDD121 pKa = 4.23LPTLNCIVAAWRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84LIRR138 pKa = 11.84SSVVWYY144 pKa = 10.5SLYY147 pKa = 10.2LTDD150 pKa = 6.14LGSSWLRR157 pKa = 11.84RR158 pKa = 11.84YY159 pKa = 6.34WTRR162 pKa = 11.84AQMIEE167 pKa = 4.04LSYY170 pKa = 11.76GCMALKK176 pKa = 10.47AMKK179 pKa = 9.93VKK181 pKa = 10.2PHH183 pKa = 5.85GLRR186 pKa = 11.84PRR188 pKa = 11.84FSKK191 pKa = 10.84DD192 pKa = 2.69GSIQGEE198 pKa = 4.45EE199 pKa = 4.08KK200 pKa = 10.43EE201 pKa = 4.3KK202 pKa = 10.42TYY204 pKa = 9.36NTSMRR209 pKa = 11.84SIWVTVYY216 pKa = 9.5STSPDD221 pKa = 2.92RR222 pKa = 11.84WKK224 pKa = 9.58TIYY227 pKa = 10.65SILYY231 pKa = 7.95YY232 pKa = 10.36KK233 pKa = 10.42KK234 pKa = 10.57LRR236 pKa = 11.84IDD238 pKa = 3.5YY239 pKa = 8.35WSSKK243 pKa = 10.09YY244 pKa = 10.7EE245 pKa = 4.52PIDD248 pKa = 3.78FNCSDD253 pKa = 3.55KK254 pKa = 11.12VHH256 pKa = 6.12VVVLSNFLPCLDD268 pKa = 4.26LEE270 pKa = 4.53YY271 pKa = 10.87NNRR274 pKa = 11.84GEE276 pKa = 4.44TVKK279 pKa = 10.84KK280 pKa = 10.05PLLSRR285 pKa = 11.84DD286 pKa = 3.15RR287 pKa = 11.84VFLINIDD294 pKa = 3.84EE295 pKa = 4.66SVCGHH300 pKa = 7.2PDD302 pKa = 3.41DD303 pKa = 5.71LKK305 pKa = 11.51SFDD308 pKa = 4.36MYY310 pKa = 11.58LEE312 pKa = 4.15
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTSLPEE24 pKa = 3.69WANYY28 pKa = 10.56LIFQEE33 pKa = 4.94EE34 pKa = 4.56EE35 pKa = 4.2CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.45VNLNAINDD55 pKa = 3.67LRR57 pKa = 11.84SCKK60 pKa = 10.1RR61 pKa = 11.84NCQMGLISKK70 pKa = 9.63HH71 pKa = 6.19AGVHH75 pKa = 5.71HH76 pKa = 6.53PAIEE80 pKa = 4.6IIAEE84 pKa = 4.07RR85 pKa = 11.84MIQEE89 pKa = 4.37STVHH93 pKa = 6.17GSSEE97 pKa = 4.18SLRR100 pKa = 11.84KK101 pKa = 9.84QGATRR106 pKa = 11.84GRR108 pKa = 11.84RR109 pKa = 11.84WNVFRR114 pKa = 11.84KK115 pKa = 6.43TQKK118 pKa = 10.89NLDD121 pKa = 4.23LPTLNCIVAAWRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84LIRR138 pKa = 11.84SSVVWYY144 pKa = 10.5SLYY147 pKa = 10.2LTDD150 pKa = 6.14LGSSWLRR157 pKa = 11.84RR158 pKa = 11.84YY159 pKa = 6.34WTRR162 pKa = 11.84AQMIEE167 pKa = 4.04LSYY170 pKa = 11.76GCMALKK176 pKa = 10.47AMKK179 pKa = 9.93VKK181 pKa = 10.2PHH183 pKa = 5.85GLRR186 pKa = 11.84PRR188 pKa = 11.84FSKK191 pKa = 10.84DD192 pKa = 2.69GSIQGEE198 pKa = 4.45EE199 pKa = 4.08KK200 pKa = 10.43EE201 pKa = 4.3KK202 pKa = 10.42TYY204 pKa = 9.36NTSMRR209 pKa = 11.84SIWVTVYY216 pKa = 9.5STSPDD221 pKa = 2.92RR222 pKa = 11.84WKK224 pKa = 9.58TIYY227 pKa = 10.65SILYY231 pKa = 7.95YY232 pKa = 10.36KK233 pKa = 10.42KK234 pKa = 10.57LRR236 pKa = 11.84IDD238 pKa = 3.5YY239 pKa = 8.35WSSKK243 pKa = 10.09YY244 pKa = 10.7EE245 pKa = 4.52PIDD248 pKa = 3.78FNCSDD253 pKa = 3.55KK254 pKa = 11.12VHH256 pKa = 6.12VVVLSNFLPCLDD268 pKa = 4.26LEE270 pKa = 4.53YY271 pKa = 10.87NNRR274 pKa = 11.84GEE276 pKa = 4.44TVKK279 pKa = 10.84KK280 pKa = 10.05PLLSRR285 pKa = 11.84DD286 pKa = 3.15RR287 pKa = 11.84VFLINIDD294 pKa = 3.84EE295 pKa = 4.66SVCGHH300 pKa = 7.2PDD302 pKa = 3.41DD303 pKa = 5.71LKK305 pKa = 11.51SFDD308 pKa = 4.36MYY310 pKa = 11.58LEE312 pKa = 4.15
Molecular weight: 36.7 kDa
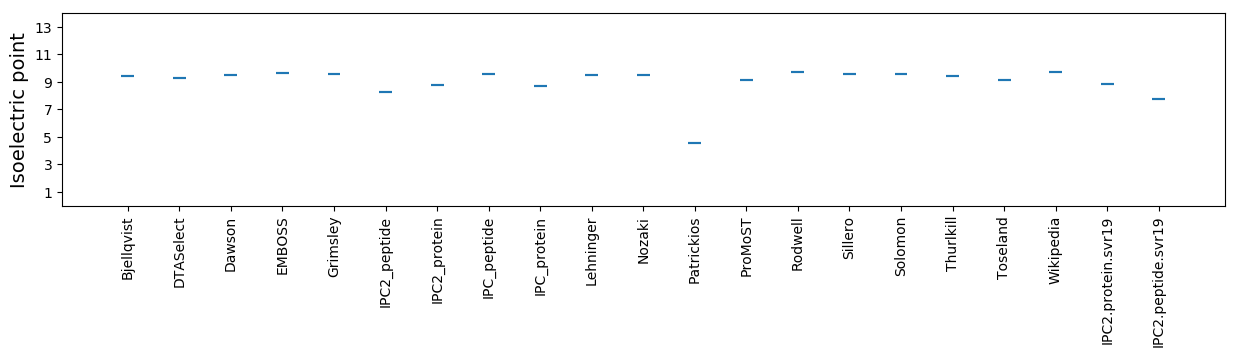
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V9I5Y2|V9I5Y2_9VIRU Rep protein OS=Papaya leaf curl alphasatellite OX=1112202 PE=4 SV=1
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTSLPEE24 pKa = 3.69WANYY28 pKa = 10.56LIFQEE33 pKa = 4.94EE34 pKa = 4.56EE35 pKa = 4.2CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.45VNLNAINDD55 pKa = 3.67LRR57 pKa = 11.84SCKK60 pKa = 10.1RR61 pKa = 11.84NCQMGLISKK70 pKa = 9.63HH71 pKa = 6.19AGVHH75 pKa = 5.71HH76 pKa = 6.53PAIEE80 pKa = 4.6IIAEE84 pKa = 4.07RR85 pKa = 11.84MIQEE89 pKa = 4.37STVHH93 pKa = 6.17GSSEE97 pKa = 4.18SLRR100 pKa = 11.84KK101 pKa = 9.84QGATRR106 pKa = 11.84GRR108 pKa = 11.84RR109 pKa = 11.84WNVFRR114 pKa = 11.84KK115 pKa = 6.43TQKK118 pKa = 10.89NLDD121 pKa = 4.23LPTLNCIVAAWRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84LIRR138 pKa = 11.84SSVVWYY144 pKa = 10.5SLYY147 pKa = 10.2LTDD150 pKa = 6.14LGSSWLRR157 pKa = 11.84RR158 pKa = 11.84YY159 pKa = 6.34WTRR162 pKa = 11.84AQMIEE167 pKa = 4.04LSYY170 pKa = 11.76GCMALKK176 pKa = 10.47AMKK179 pKa = 9.93VKK181 pKa = 10.2PHH183 pKa = 5.85GLRR186 pKa = 11.84PRR188 pKa = 11.84FSKK191 pKa = 10.84DD192 pKa = 2.69GSIQGEE198 pKa = 4.45EE199 pKa = 4.08KK200 pKa = 10.43EE201 pKa = 4.3KK202 pKa = 10.42TYY204 pKa = 9.36NTSMRR209 pKa = 11.84SIWVTVYY216 pKa = 9.5STSPDD221 pKa = 2.92RR222 pKa = 11.84WKK224 pKa = 9.58TIYY227 pKa = 10.65SILYY231 pKa = 7.95YY232 pKa = 10.36KK233 pKa = 10.42KK234 pKa = 10.57LRR236 pKa = 11.84IDD238 pKa = 3.5YY239 pKa = 8.35WSSKK243 pKa = 10.09YY244 pKa = 10.7EE245 pKa = 4.52PIDD248 pKa = 3.78FNCSDD253 pKa = 3.55KK254 pKa = 11.12VHH256 pKa = 6.12VVVLSNFLPCLDD268 pKa = 4.26LEE270 pKa = 4.53YY271 pKa = 10.87NNRR274 pKa = 11.84GEE276 pKa = 4.44TVKK279 pKa = 10.84KK280 pKa = 10.05PLLSRR285 pKa = 11.84DD286 pKa = 3.15RR287 pKa = 11.84VFLINIDD294 pKa = 3.84EE295 pKa = 4.66SVCGHH300 pKa = 7.2PDD302 pKa = 3.41DD303 pKa = 5.71LKK305 pKa = 11.51SFDD308 pKa = 4.36MYY310 pKa = 11.58LEE312 pKa = 4.15
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTSLPEE24 pKa = 3.69WANYY28 pKa = 10.56LIFQEE33 pKa = 4.94EE34 pKa = 4.56EE35 pKa = 4.2CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.45VNLNAINDD55 pKa = 3.67LRR57 pKa = 11.84SCKK60 pKa = 10.1RR61 pKa = 11.84NCQMGLISKK70 pKa = 9.63HH71 pKa = 6.19AGVHH75 pKa = 5.71HH76 pKa = 6.53PAIEE80 pKa = 4.6IIAEE84 pKa = 4.07RR85 pKa = 11.84MIQEE89 pKa = 4.37STVHH93 pKa = 6.17GSSEE97 pKa = 4.18SLRR100 pKa = 11.84KK101 pKa = 9.84QGATRR106 pKa = 11.84GRR108 pKa = 11.84RR109 pKa = 11.84WNVFRR114 pKa = 11.84KK115 pKa = 6.43TQKK118 pKa = 10.89NLDD121 pKa = 4.23LPTLNCIVAAWRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84LIRR138 pKa = 11.84SSVVWYY144 pKa = 10.5SLYY147 pKa = 10.2LTDD150 pKa = 6.14LGSSWLRR157 pKa = 11.84RR158 pKa = 11.84YY159 pKa = 6.34WTRR162 pKa = 11.84AQMIEE167 pKa = 4.04LSYY170 pKa = 11.76GCMALKK176 pKa = 10.47AMKK179 pKa = 9.93VKK181 pKa = 10.2PHH183 pKa = 5.85GLRR186 pKa = 11.84PRR188 pKa = 11.84FSKK191 pKa = 10.84DD192 pKa = 2.69GSIQGEE198 pKa = 4.45EE199 pKa = 4.08KK200 pKa = 10.43EE201 pKa = 4.3KK202 pKa = 10.42TYY204 pKa = 9.36NTSMRR209 pKa = 11.84SIWVTVYY216 pKa = 9.5STSPDD221 pKa = 2.92RR222 pKa = 11.84WKK224 pKa = 9.58TIYY227 pKa = 10.65SILYY231 pKa = 7.95YY232 pKa = 10.36KK233 pKa = 10.42KK234 pKa = 10.57LRR236 pKa = 11.84IDD238 pKa = 3.5YY239 pKa = 8.35WSSKK243 pKa = 10.09YY244 pKa = 10.7EE245 pKa = 4.52PIDD248 pKa = 3.78FNCSDD253 pKa = 3.55KK254 pKa = 11.12VHH256 pKa = 6.12VVVLSNFLPCLDD268 pKa = 4.26LEE270 pKa = 4.53YY271 pKa = 10.87NNRR274 pKa = 11.84GEE276 pKa = 4.44TVKK279 pKa = 10.84KK280 pKa = 10.05PLLSRR285 pKa = 11.84DD286 pKa = 3.15RR287 pKa = 11.84VFLINIDD294 pKa = 3.84EE295 pKa = 4.66SVCGHH300 pKa = 7.2PDD302 pKa = 3.41DD303 pKa = 5.71LKK305 pKa = 11.51SFDD308 pKa = 4.36MYY310 pKa = 11.58LEE312 pKa = 4.15
Molecular weight: 36.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
312 |
312 |
312 |
312.0 |
36.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.526 ± 0.0 | 2.885 ± 0.0 |
4.487 ± 0.0 | 5.449 ± 0.0 |
3.205 ± 0.0 | 4.167 ± 0.0 |
2.885 ± 0.0 | 6.41 ± 0.0 |
6.731 ± 0.0 | 9.615 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.564 ± 0.0 | 5.128 ± 0.0 |
3.846 ± 0.0 | 2.564 ± 0.0 |
8.013 ± 0.0 | 9.295 ± 0.0 |
5.128 ± 0.0 | 5.769 ± 0.0 |
3.205 ± 0.0 | 5.128 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
