
Prune dwarf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
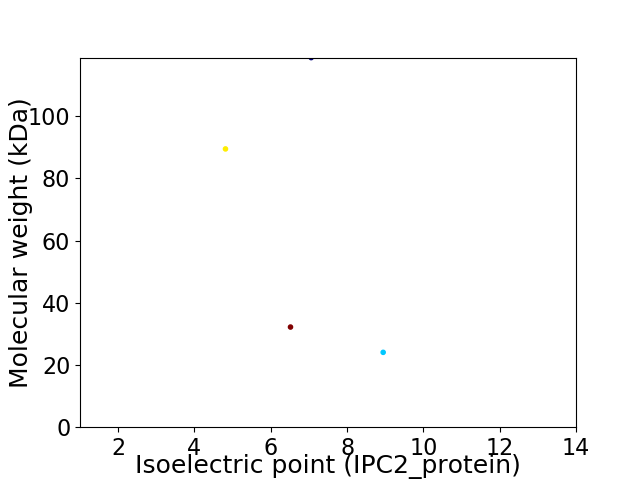
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91NQ3|Q91NQ3_9BROM RNA-directed RNA polymerase 2a OS=Prune dwarf virus OX=33760 PE=4 SV=1
MM1 pKa = 8.28DD2 pKa = 4.95SLLEE6 pKa = 3.95FLLPSHH12 pKa = 7.34DD13 pKa = 3.63EE14 pKa = 4.1DD15 pKa = 6.7AIEE18 pKa = 5.0LVDD21 pKa = 3.89TNSVKK26 pKa = 10.52PSFSLSDD33 pKa = 3.28SSGDD37 pKa = 3.14VSPIYY42 pKa = 10.33RR43 pKa = 11.84SLCKK47 pKa = 10.38FFGFLMIRR55 pKa = 11.84DD56 pKa = 3.91TVVRR60 pKa = 11.84PDD62 pKa = 3.16KK63 pKa = 10.48FTGEE67 pKa = 4.84FITLPFVIDD76 pKa = 3.83RR77 pKa = 11.84LRR79 pKa = 11.84MTFDD83 pKa = 4.83LEE85 pKa = 3.95DD86 pKa = 4.88DD87 pKa = 3.98YY88 pKa = 12.05AFPVDD93 pKa = 3.74DD94 pKa = 4.4CSYY97 pKa = 11.5DD98 pKa = 4.4LEE100 pKa = 4.49LTDD103 pKa = 4.71AQLDD107 pKa = 4.02YY108 pKa = 11.85AEE110 pKa = 5.1VLQQQKK116 pKa = 11.16DD117 pKa = 3.88FFSEE121 pKa = 4.09SLGKK125 pKa = 10.59VVVDD129 pKa = 3.62YY130 pKa = 11.29DD131 pKa = 4.14FSLDD135 pKa = 4.34SEE137 pKa = 4.71DD138 pKa = 4.42PSIKK142 pKa = 10.39DD143 pKa = 3.27VVKK146 pKa = 10.69IPDD149 pKa = 4.41EE150 pKa = 4.07ISEE153 pKa = 4.79DD154 pKa = 3.92FPQEE158 pKa = 3.89NVPAEE163 pKa = 4.1IVADD167 pKa = 3.91VPQIVSDD174 pKa = 3.88DD175 pKa = 3.79VEE177 pKa = 4.4EE178 pKa = 4.0QVVAGEE184 pKa = 5.05LIPSCEE190 pKa = 3.98TVNEE194 pKa = 4.24NVCVEE199 pKa = 4.45VKK201 pKa = 10.59FPTHH205 pKa = 6.86LAPKK209 pKa = 9.98FLATHH214 pKa = 6.71LPDD217 pKa = 3.27VDD219 pKa = 4.39RR220 pKa = 11.84VILVDD225 pKa = 3.76GDD227 pKa = 4.1TVDD230 pKa = 5.55GFDD233 pKa = 5.0KK234 pKa = 11.09LISRR238 pKa = 11.84NDD240 pKa = 2.86WFFLKK245 pKa = 10.5KK246 pKa = 10.3LLPNIPLPSDD256 pKa = 3.51GDD258 pKa = 3.89VLLSGIKK265 pKa = 8.34PTSPGVLQFALDD277 pKa = 3.69EE278 pKa = 4.47LFPYY282 pKa = 10.28HH283 pKa = 6.73NLVDD287 pKa = 3.99DD288 pKa = 4.58RR289 pKa = 11.84FFQEE293 pKa = 5.27LVEE296 pKa = 4.7CNDD299 pKa = 3.13IALEE303 pKa = 4.58LDD305 pKa = 3.8DD306 pKa = 5.99CSFDD310 pKa = 3.71LSHH313 pKa = 6.66FVSWDD318 pKa = 3.14KK319 pKa = 11.42GKK321 pKa = 11.01SGVDD325 pKa = 3.33STLMTGQTSNRR336 pKa = 11.84QSTFRR341 pKa = 11.84EE342 pKa = 3.88VALSVKK348 pKa = 9.06KK349 pKa = 10.68RR350 pKa = 11.84NMNVPILNTVLNVEE364 pKa = 4.36DD365 pKa = 4.03VSNKK369 pKa = 8.55IVNRR373 pKa = 11.84FFEE376 pKa = 4.53TVVDD380 pKa = 4.07INLLTTLPDD389 pKa = 3.63VISIGEE395 pKa = 3.9VNYY398 pKa = 10.44FSDD401 pKa = 4.0YY402 pKa = 11.43LKK404 pKa = 11.18GKK406 pKa = 9.97AINDD410 pKa = 3.53DD411 pKa = 3.92EE412 pKa = 6.18LYY414 pKa = 10.87VDD416 pKa = 5.22PICLVSMDD424 pKa = 4.85KK425 pKa = 10.07YY426 pKa = 10.41RR427 pKa = 11.84HH428 pKa = 5.64MIKK431 pKa = 10.09SQLKK435 pKa = 8.41PVEE438 pKa = 4.72DD439 pKa = 3.54NSLMFEE445 pKa = 4.3RR446 pKa = 11.84PLAATITFHH455 pKa = 7.97DD456 pKa = 4.18KK457 pKa = 10.93GKK459 pKa = 10.93VMSTSPIFLAAATRR473 pKa = 11.84LFACLNEE480 pKa = 4.85KK481 pKa = 10.33ISIPSGKK488 pKa = 9.3YY489 pKa = 8.95HH490 pKa = 6.71QLFSLDD496 pKa = 3.33AEE498 pKa = 4.52RR499 pKa = 11.84FDD501 pKa = 5.45SVRR504 pKa = 11.84FWKK507 pKa = 10.54EE508 pKa = 3.1VDD510 pKa = 3.75FSKK513 pKa = 10.67FDD515 pKa = 3.3KK516 pKa = 10.83SQQEE520 pKa = 4.0LHH522 pKa = 6.95HH523 pKa = 6.46EE524 pKa = 4.12IQRR527 pKa = 11.84KK528 pKa = 8.29IFLRR532 pKa = 11.84LGVPQEE538 pKa = 4.65FVNTWFTSHH547 pKa = 6.31CRR549 pKa = 11.84SHH551 pKa = 8.38ISDD554 pKa = 3.32ASGLRR559 pKa = 11.84FSVNYY564 pKa = 8.2QRR566 pKa = 11.84RR567 pKa = 11.84AGDD570 pKa = 3.01ACTYY574 pKa = 10.2LGNTIVTLAALCYY587 pKa = 10.4VYY589 pKa = 10.82DD590 pKa = 3.94LRR592 pKa = 11.84SSNVAMVVASGDD604 pKa = 3.49DD605 pKa = 3.69SLIGSYY611 pKa = 10.72SEE613 pKa = 4.19LDD615 pKa = 3.0RR616 pKa = 11.84SFEE619 pKa = 4.32HH620 pKa = 7.2LFSTLFNFEE629 pKa = 4.2AKK631 pKa = 10.1FPHH634 pKa = 5.93NQPFICSKK642 pKa = 10.66FLLTMPTKK650 pKa = 10.73GGGKK654 pKa = 9.22QVVAVPNPAKK664 pKa = 10.68LLIKK668 pKa = 10.57LGVKK672 pKa = 10.41SMTVDD677 pKa = 4.69KK678 pKa = 10.9FDD680 pKa = 5.55DD681 pKa = 3.94WFQSWLDD688 pKa = 3.97LIHH691 pKa = 6.63YY692 pKa = 9.88FDD694 pKa = 5.6DD695 pKa = 4.5YY696 pKa = 11.83YY697 pKa = 11.18LIEE700 pKa = 4.63VVCSMTSYY708 pKa = 10.81RR709 pKa = 11.84YY710 pKa = 9.59IRR712 pKa = 11.84RR713 pKa = 11.84PSQFLVSAISSLKK726 pKa = 10.88SLFANKK732 pKa = 9.63KK733 pKa = 7.62KK734 pKa = 10.96CKK736 pKa = 10.24DD737 pKa = 3.38FLFPGLRR744 pKa = 11.84DD745 pKa = 3.79KK746 pKa = 11.67NPDD749 pKa = 2.76LSMRR753 pKa = 11.84LDD755 pKa = 3.65TPIKK759 pKa = 10.41IRR761 pKa = 11.84DD762 pKa = 3.65VTSKK766 pKa = 9.59RR767 pKa = 11.84TRR769 pKa = 11.84GKK771 pKa = 10.83KK772 pKa = 9.36KK773 pKa = 9.52MKK775 pKa = 9.9EE776 pKa = 3.82NSHH779 pKa = 5.55FQIDD783 pKa = 4.18GKK785 pKa = 11.4VGGG788 pKa = 4.24
MM1 pKa = 8.28DD2 pKa = 4.95SLLEE6 pKa = 3.95FLLPSHH12 pKa = 7.34DD13 pKa = 3.63EE14 pKa = 4.1DD15 pKa = 6.7AIEE18 pKa = 5.0LVDD21 pKa = 3.89TNSVKK26 pKa = 10.52PSFSLSDD33 pKa = 3.28SSGDD37 pKa = 3.14VSPIYY42 pKa = 10.33RR43 pKa = 11.84SLCKK47 pKa = 10.38FFGFLMIRR55 pKa = 11.84DD56 pKa = 3.91TVVRR60 pKa = 11.84PDD62 pKa = 3.16KK63 pKa = 10.48FTGEE67 pKa = 4.84FITLPFVIDD76 pKa = 3.83RR77 pKa = 11.84LRR79 pKa = 11.84MTFDD83 pKa = 4.83LEE85 pKa = 3.95DD86 pKa = 4.88DD87 pKa = 3.98YY88 pKa = 12.05AFPVDD93 pKa = 3.74DD94 pKa = 4.4CSYY97 pKa = 11.5DD98 pKa = 4.4LEE100 pKa = 4.49LTDD103 pKa = 4.71AQLDD107 pKa = 4.02YY108 pKa = 11.85AEE110 pKa = 5.1VLQQQKK116 pKa = 11.16DD117 pKa = 3.88FFSEE121 pKa = 4.09SLGKK125 pKa = 10.59VVVDD129 pKa = 3.62YY130 pKa = 11.29DD131 pKa = 4.14FSLDD135 pKa = 4.34SEE137 pKa = 4.71DD138 pKa = 4.42PSIKK142 pKa = 10.39DD143 pKa = 3.27VVKK146 pKa = 10.69IPDD149 pKa = 4.41EE150 pKa = 4.07ISEE153 pKa = 4.79DD154 pKa = 3.92FPQEE158 pKa = 3.89NVPAEE163 pKa = 4.1IVADD167 pKa = 3.91VPQIVSDD174 pKa = 3.88DD175 pKa = 3.79VEE177 pKa = 4.4EE178 pKa = 4.0QVVAGEE184 pKa = 5.05LIPSCEE190 pKa = 3.98TVNEE194 pKa = 4.24NVCVEE199 pKa = 4.45VKK201 pKa = 10.59FPTHH205 pKa = 6.86LAPKK209 pKa = 9.98FLATHH214 pKa = 6.71LPDD217 pKa = 3.27VDD219 pKa = 4.39RR220 pKa = 11.84VILVDD225 pKa = 3.76GDD227 pKa = 4.1TVDD230 pKa = 5.55GFDD233 pKa = 5.0KK234 pKa = 11.09LISRR238 pKa = 11.84NDD240 pKa = 2.86WFFLKK245 pKa = 10.5KK246 pKa = 10.3LLPNIPLPSDD256 pKa = 3.51GDD258 pKa = 3.89VLLSGIKK265 pKa = 8.34PTSPGVLQFALDD277 pKa = 3.69EE278 pKa = 4.47LFPYY282 pKa = 10.28HH283 pKa = 6.73NLVDD287 pKa = 3.99DD288 pKa = 4.58RR289 pKa = 11.84FFQEE293 pKa = 5.27LVEE296 pKa = 4.7CNDD299 pKa = 3.13IALEE303 pKa = 4.58LDD305 pKa = 3.8DD306 pKa = 5.99CSFDD310 pKa = 3.71LSHH313 pKa = 6.66FVSWDD318 pKa = 3.14KK319 pKa = 11.42GKK321 pKa = 11.01SGVDD325 pKa = 3.33STLMTGQTSNRR336 pKa = 11.84QSTFRR341 pKa = 11.84EE342 pKa = 3.88VALSVKK348 pKa = 9.06KK349 pKa = 10.68RR350 pKa = 11.84NMNVPILNTVLNVEE364 pKa = 4.36DD365 pKa = 4.03VSNKK369 pKa = 8.55IVNRR373 pKa = 11.84FFEE376 pKa = 4.53TVVDD380 pKa = 4.07INLLTTLPDD389 pKa = 3.63VISIGEE395 pKa = 3.9VNYY398 pKa = 10.44FSDD401 pKa = 4.0YY402 pKa = 11.43LKK404 pKa = 11.18GKK406 pKa = 9.97AINDD410 pKa = 3.53DD411 pKa = 3.92EE412 pKa = 6.18LYY414 pKa = 10.87VDD416 pKa = 5.22PICLVSMDD424 pKa = 4.85KK425 pKa = 10.07YY426 pKa = 10.41RR427 pKa = 11.84HH428 pKa = 5.64MIKK431 pKa = 10.09SQLKK435 pKa = 8.41PVEE438 pKa = 4.72DD439 pKa = 3.54NSLMFEE445 pKa = 4.3RR446 pKa = 11.84PLAATITFHH455 pKa = 7.97DD456 pKa = 4.18KK457 pKa = 10.93GKK459 pKa = 10.93VMSTSPIFLAAATRR473 pKa = 11.84LFACLNEE480 pKa = 4.85KK481 pKa = 10.33ISIPSGKK488 pKa = 9.3YY489 pKa = 8.95HH490 pKa = 6.71QLFSLDD496 pKa = 3.33AEE498 pKa = 4.52RR499 pKa = 11.84FDD501 pKa = 5.45SVRR504 pKa = 11.84FWKK507 pKa = 10.54EE508 pKa = 3.1VDD510 pKa = 3.75FSKK513 pKa = 10.67FDD515 pKa = 3.3KK516 pKa = 10.83SQQEE520 pKa = 4.0LHH522 pKa = 6.95HH523 pKa = 6.46EE524 pKa = 4.12IQRR527 pKa = 11.84KK528 pKa = 8.29IFLRR532 pKa = 11.84LGVPQEE538 pKa = 4.65FVNTWFTSHH547 pKa = 6.31CRR549 pKa = 11.84SHH551 pKa = 8.38ISDD554 pKa = 3.32ASGLRR559 pKa = 11.84FSVNYY564 pKa = 8.2QRR566 pKa = 11.84RR567 pKa = 11.84AGDD570 pKa = 3.01ACTYY574 pKa = 10.2LGNTIVTLAALCYY587 pKa = 10.4VYY589 pKa = 10.82DD590 pKa = 3.94LRR592 pKa = 11.84SSNVAMVVASGDD604 pKa = 3.49DD605 pKa = 3.69SLIGSYY611 pKa = 10.72SEE613 pKa = 4.19LDD615 pKa = 3.0RR616 pKa = 11.84SFEE619 pKa = 4.32HH620 pKa = 7.2LFSTLFNFEE629 pKa = 4.2AKK631 pKa = 10.1FPHH634 pKa = 5.93NQPFICSKK642 pKa = 10.66FLLTMPTKK650 pKa = 10.73GGGKK654 pKa = 9.22QVVAVPNPAKK664 pKa = 10.68LLIKK668 pKa = 10.57LGVKK672 pKa = 10.41SMTVDD677 pKa = 4.69KK678 pKa = 10.9FDD680 pKa = 5.55DD681 pKa = 3.94WFQSWLDD688 pKa = 3.97LIHH691 pKa = 6.63YY692 pKa = 9.88FDD694 pKa = 5.6DD695 pKa = 4.5YY696 pKa = 11.83YY697 pKa = 11.18LIEE700 pKa = 4.63VVCSMTSYY708 pKa = 10.81RR709 pKa = 11.84YY710 pKa = 9.59IRR712 pKa = 11.84RR713 pKa = 11.84PSQFLVSAISSLKK726 pKa = 10.88SLFANKK732 pKa = 9.63KK733 pKa = 7.62KK734 pKa = 10.96CKK736 pKa = 10.24DD737 pKa = 3.38FLFPGLRR744 pKa = 11.84DD745 pKa = 3.79KK746 pKa = 11.67NPDD749 pKa = 2.76LSMRR753 pKa = 11.84LDD755 pKa = 3.65TPIKK759 pKa = 10.41IRR761 pKa = 11.84DD762 pKa = 3.65VTSKK766 pKa = 9.59RR767 pKa = 11.84TRR769 pKa = 11.84GKK771 pKa = 10.83KK772 pKa = 9.36KK773 pKa = 9.52MKK775 pKa = 9.9EE776 pKa = 3.82NSHH779 pKa = 5.55FQIDD783 pKa = 4.18GKK785 pKa = 11.4VGGG788 pKa = 4.24
Molecular weight: 89.5 kDa
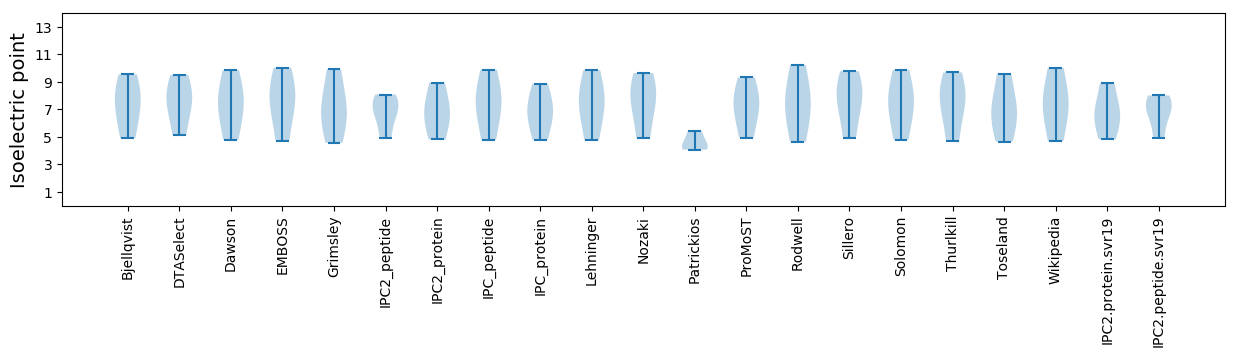
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91NQ3|Q91NQ3_9BROM RNA-directed RNA polymerase 2a OS=Prune dwarf virus OX=33760 PE=4 SV=1
MM1 pKa = 7.37SGKK4 pKa = 10.02AIKK7 pKa = 10.06SGKK10 pKa = 7.27PTTRR14 pKa = 11.84SQSFALARR22 pKa = 11.84KK23 pKa = 9.26NNNTTPPAGFVKK35 pKa = 10.47KK36 pKa = 9.7QFPSGSSKK44 pKa = 10.92SISEE48 pKa = 4.12WMLHH52 pKa = 5.59GPNVPVKK59 pKa = 9.8SFSGMISRR67 pKa = 11.84TEE69 pKa = 3.77NLTVNSTASGVYY81 pKa = 7.41YY82 pKa = 9.7TMKK85 pKa = 10.08VRR87 pKa = 11.84EE88 pKa = 4.32LFKK91 pKa = 11.24DD92 pKa = 3.87FAVDD96 pKa = 3.3TKK98 pKa = 11.17VYY100 pKa = 10.39GIVFRR105 pKa = 11.84YY106 pKa = 9.74CLDD109 pKa = 3.27VSNGVYY115 pKa = 10.84GLIKK119 pKa = 10.62GFDD122 pKa = 3.58VNAPVAPNPLQRR134 pKa = 11.84RR135 pKa = 11.84KK136 pKa = 8.55FTAKK140 pKa = 9.56QASGVQILAPTGMTVGDD157 pKa = 4.48IPDD160 pKa = 4.59DD161 pKa = 3.27LWFVIKK167 pKa = 10.42YY168 pKa = 10.8DD169 pKa = 3.69NAFQPNVPVWFCTQYY184 pKa = 10.72LQHH187 pKa = 6.99SMPKK191 pKa = 9.55RR192 pKa = 11.84VEE194 pKa = 4.13VPDD197 pKa = 3.55SVLYY201 pKa = 10.74AEE203 pKa = 5.5RR204 pKa = 11.84DD205 pKa = 3.63TALMDD210 pKa = 3.94AMDD213 pKa = 5.32KK214 pKa = 10.57IVSGG218 pKa = 3.91
MM1 pKa = 7.37SGKK4 pKa = 10.02AIKK7 pKa = 10.06SGKK10 pKa = 7.27PTTRR14 pKa = 11.84SQSFALARR22 pKa = 11.84KK23 pKa = 9.26NNNTTPPAGFVKK35 pKa = 10.47KK36 pKa = 9.7QFPSGSSKK44 pKa = 10.92SISEE48 pKa = 4.12WMLHH52 pKa = 5.59GPNVPVKK59 pKa = 9.8SFSGMISRR67 pKa = 11.84TEE69 pKa = 3.77NLTVNSTASGVYY81 pKa = 7.41YY82 pKa = 9.7TMKK85 pKa = 10.08VRR87 pKa = 11.84EE88 pKa = 4.32LFKK91 pKa = 11.24DD92 pKa = 3.87FAVDD96 pKa = 3.3TKK98 pKa = 11.17VYY100 pKa = 10.39GIVFRR105 pKa = 11.84YY106 pKa = 9.74CLDD109 pKa = 3.27VSNGVYY115 pKa = 10.84GLIKK119 pKa = 10.62GFDD122 pKa = 3.58VNAPVAPNPLQRR134 pKa = 11.84RR135 pKa = 11.84KK136 pKa = 8.55FTAKK140 pKa = 9.56QASGVQILAPTGMTVGDD157 pKa = 4.48IPDD160 pKa = 4.59DD161 pKa = 3.27LWFVIKK167 pKa = 10.42YY168 pKa = 10.8DD169 pKa = 3.69NAFQPNVPVWFCTQYY184 pKa = 10.72LQHH187 pKa = 6.99SMPKK191 pKa = 9.55RR192 pKa = 11.84VEE194 pKa = 4.13VPDD197 pKa = 3.55SVLYY201 pKa = 10.74AEE203 pKa = 5.5RR204 pKa = 11.84DD205 pKa = 3.63TALMDD210 pKa = 3.94AMDD213 pKa = 5.32KK214 pKa = 10.57IVSGG218 pKa = 3.91
Molecular weight: 24.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2354 |
218 |
1055 |
588.5 |
66.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.202 ± 0.894 | 1.997 ± 0.416 |
7.562 ± 0.917 | 5.353 ± 0.428 |
5.183 ± 0.919 | 4.97 ± 0.348 |
2.421 ± 0.376 | 5.65 ± 0.314 |
7.604 ± 0.581 | 8.794 ± 0.617 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.202 | 3.526 ± 0.273 |
4.291 ± 0.538 | 2.889 ± 0.175 |
4.418 ± 0.428 | 8.709 ± 0.468 |
5.692 ± 0.495 | 8.156 ± 0.452 |
0.892 ± 0.065 | 3.441 ± 0.473 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
