
Pelagibacter phage HTVC031P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Pelagivirus; unclassified Pelagivirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
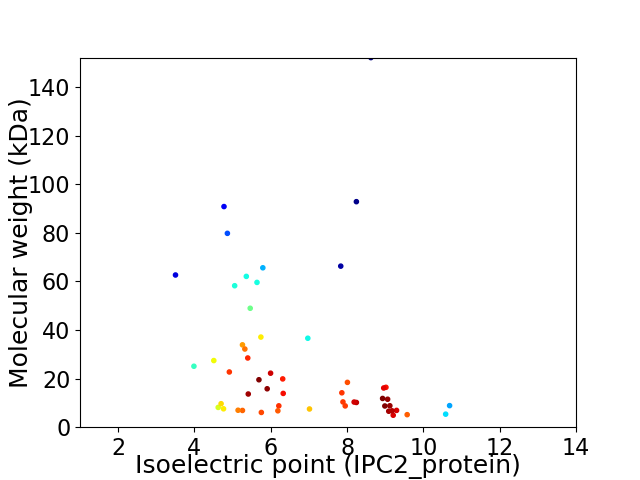
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NU04|A0A4Y1NU04_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC031P OX=2283017 GN=P031_gp36 PE=4 SV=1
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.28TGNGTTTTYY17 pKa = 10.69AIPFSYY23 pKa = 10.37RR24 pKa = 11.84SAEE27 pKa = 4.12DD28 pKa = 3.34LSTTVAGVNVTAYY41 pKa = 9.24TLDD44 pKa = 3.52AAGTNLTFTTAPANNAAIEE63 pKa = 4.04IRR65 pKa = 11.84RR66 pKa = 11.84TTSQNTKK73 pKa = 10.27LVDD76 pKa = 3.75YY77 pKa = 10.91VSGSVLTEE85 pKa = 3.67NDD87 pKa = 3.94LDD89 pKa = 4.06TDD91 pKa = 3.89SDD93 pKa = 3.77QAFYY97 pKa = 10.38MSQEE101 pKa = 4.76AIDD104 pKa = 3.77KK105 pKa = 10.49AGDD108 pKa = 3.78VISLDD113 pKa = 3.33NVDD116 pKa = 5.31FNWDD120 pKa = 3.32IQNKK124 pKa = 8.13RR125 pKa = 11.84LKK127 pKa = 10.59NVADD131 pKa = 3.99PVDD134 pKa = 3.52NTDD137 pKa = 3.33AVNKK141 pKa = 9.89QFISTNIPNITTVAGIASDD160 pKa = 3.7VTTVANNNANITAVAADD177 pKa = 3.65ATDD180 pKa = 3.18IGTVATNISSVNTVATNINDD200 pKa = 4.03VIAVANDD207 pKa = 3.18LAEE210 pKa = 4.27AVSEE214 pKa = 4.53VEE216 pKa = 4.57TVANDD221 pKa = 3.58LNEE224 pKa = 4.27ATSEE228 pKa = 3.92IDD230 pKa = 3.42TVATNITNVNTVGTNIANVNTVAGVNANVTTVAGISADD268 pKa = 3.56VTGVAGISTAVTNVNSNSTNINAVNANSANINTVAGISSDD308 pKa = 3.61VTSVANISSDD318 pKa = 3.3VAAVEE323 pKa = 4.67NIASNVTTVAGISSDD338 pKa = 3.67VTSVAGISSNVTTVASNNANVSAVGGAITNVNNVGNSIASVNTVATNLASVNSFANTYY396 pKa = 10.15LGASATAPTQDD407 pKa = 4.04PDD409 pKa = 4.31GSSLDD414 pKa = 4.3LGDD417 pKa = 5.38LYY419 pKa = 11.35FDD421 pKa = 3.82TASNTMKK428 pKa = 10.62VYY430 pKa = 10.66GSSGWTAAGSSVNGTASRR448 pKa = 11.84FKK450 pKa = 9.8YY451 pKa = 8.75TATSGQTTFTGTDD464 pKa = 3.49DD465 pKa = 3.89NSATLAYY472 pKa = 10.09DD473 pKa = 3.52AGFLDD478 pKa = 4.4VYY480 pKa = 11.11LNGIRR485 pKa = 11.84LVNGTDD491 pKa = 4.05FTASTGNSIVLTTGANLNDD510 pKa = 3.44ILEE513 pKa = 4.34IVAFGTFALANFSITDD529 pKa = 3.59ATDD532 pKa = 3.24VPPLGSAGQALVVNSGGTALEE553 pKa = 4.1FSNASSAEE561 pKa = 3.88VYY563 pKa = 10.55GFHH566 pKa = 7.42KK567 pKa = 10.63DD568 pKa = 3.38SNGDD572 pKa = 4.12LIVTTTNQGVDD583 pKa = 3.74NISSATYY590 pKa = 8.63ATFDD594 pKa = 3.88DD595 pKa = 4.41VLFSASGFTFSLNNGEE611 pKa = 4.99LIATII616 pKa = 4.25
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 7.28TGNGTTTTYY17 pKa = 10.69AIPFSYY23 pKa = 10.37RR24 pKa = 11.84SAEE27 pKa = 4.12DD28 pKa = 3.34LSTTVAGVNVTAYY41 pKa = 9.24TLDD44 pKa = 3.52AAGTNLTFTTAPANNAAIEE63 pKa = 4.04IRR65 pKa = 11.84RR66 pKa = 11.84TTSQNTKK73 pKa = 10.27LVDD76 pKa = 3.75YY77 pKa = 10.91VSGSVLTEE85 pKa = 3.67NDD87 pKa = 3.94LDD89 pKa = 4.06TDD91 pKa = 3.89SDD93 pKa = 3.77QAFYY97 pKa = 10.38MSQEE101 pKa = 4.76AIDD104 pKa = 3.77KK105 pKa = 10.49AGDD108 pKa = 3.78VISLDD113 pKa = 3.33NVDD116 pKa = 5.31FNWDD120 pKa = 3.32IQNKK124 pKa = 8.13RR125 pKa = 11.84LKK127 pKa = 10.59NVADD131 pKa = 3.99PVDD134 pKa = 3.52NTDD137 pKa = 3.33AVNKK141 pKa = 9.89QFISTNIPNITTVAGIASDD160 pKa = 3.7VTTVANNNANITAVAADD177 pKa = 3.65ATDD180 pKa = 3.18IGTVATNISSVNTVATNINDD200 pKa = 4.03VIAVANDD207 pKa = 3.18LAEE210 pKa = 4.27AVSEE214 pKa = 4.53VEE216 pKa = 4.57TVANDD221 pKa = 3.58LNEE224 pKa = 4.27ATSEE228 pKa = 3.92IDD230 pKa = 3.42TVATNITNVNTVGTNIANVNTVAGVNANVTTVAGISADD268 pKa = 3.56VTGVAGISTAVTNVNSNSTNINAVNANSANINTVAGISSDD308 pKa = 3.61VTSVANISSDD318 pKa = 3.3VAAVEE323 pKa = 4.67NIASNVTTVAGISSDD338 pKa = 3.67VTSVAGISSNVTTVASNNANVSAVGGAITNVNNVGNSIASVNTVATNLASVNSFANTYY396 pKa = 10.15LGASATAPTQDD407 pKa = 4.04PDD409 pKa = 4.31GSSLDD414 pKa = 4.3LGDD417 pKa = 5.38LYY419 pKa = 11.35FDD421 pKa = 3.82TASNTMKK428 pKa = 10.62VYY430 pKa = 10.66GSSGWTAAGSSVNGTASRR448 pKa = 11.84FKK450 pKa = 9.8YY451 pKa = 8.75TATSGQTTFTGTDD464 pKa = 3.49DD465 pKa = 3.89NSATLAYY472 pKa = 10.09DD473 pKa = 3.52AGFLDD478 pKa = 4.4VYY480 pKa = 11.11LNGIRR485 pKa = 11.84LVNGTDD491 pKa = 4.05FTASTGNSIVLTTGANLNDD510 pKa = 3.44ILEE513 pKa = 4.34IVAFGTFALANFSITDD529 pKa = 3.59ATDD532 pKa = 3.24VPPLGSAGQALVVNSGGTALEE553 pKa = 4.1FSNASSAEE561 pKa = 3.88VYY563 pKa = 10.55GFHH566 pKa = 7.42KK567 pKa = 10.63DD568 pKa = 3.38SNGDD572 pKa = 4.12LIVTTTNQGVDD583 pKa = 3.74NISSATYY590 pKa = 8.63ATFDD594 pKa = 3.88DD595 pKa = 4.41VLFSASGFTFSLNNGEE611 pKa = 4.99LIATII616 pKa = 4.25
Molecular weight: 62.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NTZ6|A0A4Y1NTZ6_9CAUD Lectin domain containing protein OS=Pelagibacter phage HTVC031P OX=2283017 GN=P031_gp42 PE=4 SV=1
MM1 pKa = 7.28PAKK4 pKa = 10.22RR5 pKa = 11.84YY6 pKa = 8.18QSPSGGLNAAGRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.67FKK22 pKa = 10.8RR23 pKa = 11.84KK24 pKa = 7.88TGANLKK30 pKa = 10.69APVTGKK36 pKa = 10.18VKK38 pKa = 10.54RR39 pKa = 11.84GSKK42 pKa = 8.47AAKK45 pKa = 9.57RR46 pKa = 11.84RR47 pKa = 11.84ASFCARR53 pKa = 11.84MSGVKK58 pKa = 10.22GAMKK62 pKa = 10.17KK63 pKa = 10.48PNGQPTRR70 pKa = 11.84KK71 pKa = 9.87ALALRR76 pKa = 11.84KK77 pKa = 8.38WKK79 pKa = 10.43CRR81 pKa = 3.49
MM1 pKa = 7.28PAKK4 pKa = 10.22RR5 pKa = 11.84YY6 pKa = 8.18QSPSGGLNAAGRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.67FKK22 pKa = 10.8RR23 pKa = 11.84KK24 pKa = 7.88TGANLKK30 pKa = 10.69APVTGKK36 pKa = 10.18VKK38 pKa = 10.54RR39 pKa = 11.84GSKK42 pKa = 8.47AAKK45 pKa = 9.57RR46 pKa = 11.84RR47 pKa = 11.84ASFCARR53 pKa = 11.84MSGVKK58 pKa = 10.22GAMKK62 pKa = 10.17KK63 pKa = 10.48PNGQPTRR70 pKa = 11.84KK71 pKa = 9.87ALALRR76 pKa = 11.84KK77 pKa = 8.38WKK79 pKa = 10.43CRR81 pKa = 3.49
Molecular weight: 8.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12937 |
40 |
1345 |
244.1 |
27.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.668 ± 0.421 | 0.819 ± 0.15 |
6.068 ± 0.212 | 6.292 ± 0.429 |
3.795 ± 0.149 | 6.423 ± 0.373 |
1.639 ± 0.187 | 6.23 ± 0.184 |
8.333 ± 0.692 | 7.969 ± 0.336 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.373 ± 0.189 | 6.47 ± 0.427 |
3.115 ± 0.202 | 4.019 ± 0.265 |
3.594 ± 0.249 | 6.671 ± 0.445 |
7.49 ± 0.552 | 6.137 ± 0.389 |
1.19 ± 0.114 | 3.703 ± 0.19 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
