
Rotavirus D chicken/05V0049/DEU/2005
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus D
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
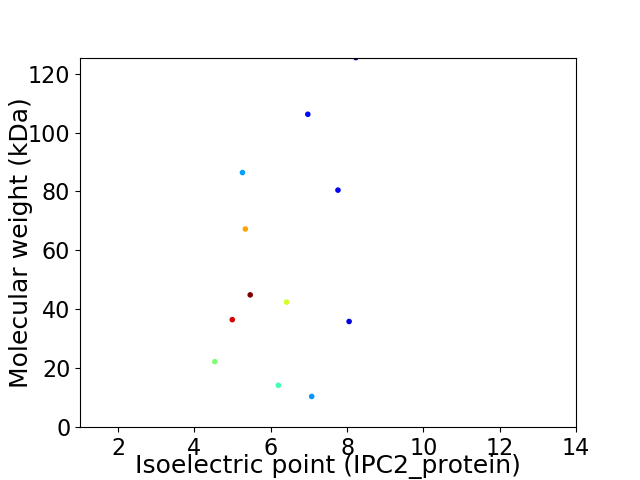
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
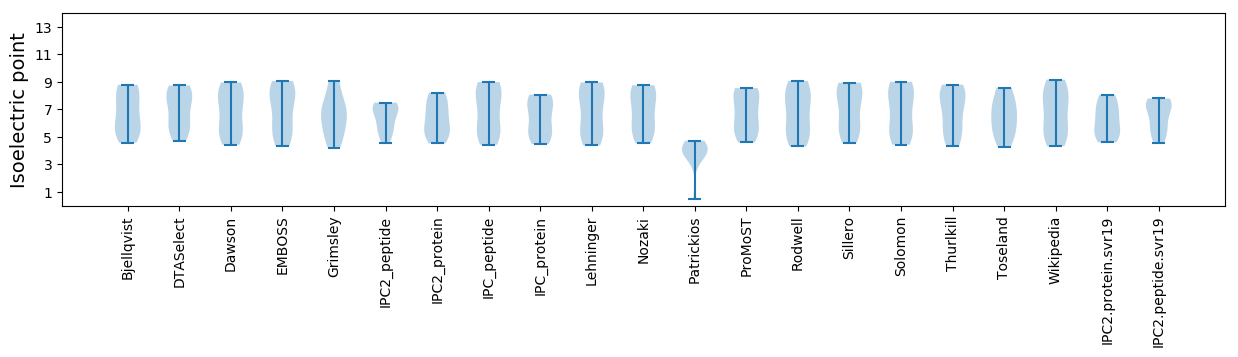
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2EBV0|E2EBV0_9REOV NSP5 OS=Rotavirus D chicken/05V0049/DEU/2005 OX=884200 PE=4 SV=1
MM1 pKa = 7.57MDD3 pKa = 4.62DD4 pKa = 4.51LDD6 pKa = 5.88FNFEE10 pKa = 4.51SNLPEE15 pKa = 4.44ISLISSRR22 pKa = 11.84AGTTYY27 pKa = 10.73TKK29 pKa = 9.8IDD31 pKa = 3.75YY32 pKa = 10.86DD33 pKa = 4.1EE34 pKa = 6.2DD35 pKa = 3.73MLLDD39 pKa = 5.8DD40 pKa = 4.82ITPSDD45 pKa = 3.88SASSQDD51 pKa = 3.19TNQRR55 pKa = 11.84TFRR58 pKa = 11.84EE59 pKa = 4.21KK60 pKa = 10.59SFKK63 pKa = 10.14SSSMVSQCDD72 pKa = 3.23EE73 pKa = 4.67DD74 pKa = 6.68DD75 pKa = 3.77IASQEE80 pKa = 4.1MNKK83 pKa = 10.39LEE85 pKa = 4.42TIVNSACADD94 pKa = 3.62EE95 pKa = 4.4QQNIDD100 pKa = 3.74DD101 pKa = 3.81WNEE104 pKa = 3.71YY105 pKa = 10.71LEE107 pKa = 4.55EE108 pKa = 4.14NSGIKK113 pKa = 9.89IMEE116 pKa = 4.53GKK118 pKa = 10.55VSTNEE123 pKa = 3.27VDD125 pKa = 3.57LNGVFEE131 pKa = 4.53SKK133 pKa = 10.22ILNRR137 pKa = 11.84NSIINKK143 pKa = 9.71DD144 pKa = 4.03NIDD147 pKa = 3.65SAVKK151 pKa = 10.32KK152 pKa = 10.01KK153 pKa = 11.21ANINKK158 pKa = 8.62MNMHH162 pKa = 6.67DD163 pKa = 3.49TSSDD167 pKa = 3.54EE168 pKa = 4.1EE169 pKa = 4.46CNRR172 pKa = 11.84NCKK175 pKa = 9.73CCKK178 pKa = 9.85KK179 pKa = 10.18LRR181 pKa = 11.84KK182 pKa = 9.04LRR184 pKa = 11.84KK185 pKa = 9.46RR186 pKa = 11.84MSILIAEE193 pKa = 4.76SYY195 pKa = 10.62
MM1 pKa = 7.57MDD3 pKa = 4.62DD4 pKa = 4.51LDD6 pKa = 5.88FNFEE10 pKa = 4.51SNLPEE15 pKa = 4.44ISLISSRR22 pKa = 11.84AGTTYY27 pKa = 10.73TKK29 pKa = 9.8IDD31 pKa = 3.75YY32 pKa = 10.86DD33 pKa = 4.1EE34 pKa = 6.2DD35 pKa = 3.73MLLDD39 pKa = 5.8DD40 pKa = 4.82ITPSDD45 pKa = 3.88SASSQDD51 pKa = 3.19TNQRR55 pKa = 11.84TFRR58 pKa = 11.84EE59 pKa = 4.21KK60 pKa = 10.59SFKK63 pKa = 10.14SSSMVSQCDD72 pKa = 3.23EE73 pKa = 4.67DD74 pKa = 6.68DD75 pKa = 3.77IASQEE80 pKa = 4.1MNKK83 pKa = 10.39LEE85 pKa = 4.42TIVNSACADD94 pKa = 3.62EE95 pKa = 4.4QQNIDD100 pKa = 3.74DD101 pKa = 3.81WNEE104 pKa = 3.71YY105 pKa = 10.71LEE107 pKa = 4.55EE108 pKa = 4.14NSGIKK113 pKa = 9.89IMEE116 pKa = 4.53GKK118 pKa = 10.55VSTNEE123 pKa = 3.27VDD125 pKa = 3.57LNGVFEE131 pKa = 4.53SKK133 pKa = 10.22ILNRR137 pKa = 11.84NSIINKK143 pKa = 9.71DD144 pKa = 4.03NIDD147 pKa = 3.65SAVKK151 pKa = 10.32KK152 pKa = 10.01KK153 pKa = 11.21ANINKK158 pKa = 8.62MNMHH162 pKa = 6.67DD163 pKa = 3.49TSSDD167 pKa = 3.54EE168 pKa = 4.1EE169 pKa = 4.46CNRR172 pKa = 11.84NCKK175 pKa = 9.73CCKK178 pKa = 9.85KK179 pKa = 10.18LRR181 pKa = 11.84KK182 pKa = 9.04LRR184 pKa = 11.84KK185 pKa = 9.46RR186 pKa = 11.84MSILIAEE193 pKa = 4.76SYY195 pKa = 10.62
Molecular weight: 22.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2EBU0|E2EBU0_9REOV Inner capsid protein VP2 OS=Rotavirus D chicken/05V0049/DEU/2005 OX=884200 PE=3 SV=1
MM1 pKa = 7.01GTYY4 pKa = 10.78NLLLQKK10 pKa = 9.6YY11 pKa = 8.78LRR13 pKa = 11.84WLYY16 pKa = 11.11GKK18 pKa = 9.58DD19 pKa = 2.94ATVIPIFITNDD30 pKa = 3.14QNNQEE35 pKa = 4.26ILNSIVDD42 pKa = 3.79YY43 pKa = 11.04AKK45 pKa = 10.61QLAIDD50 pKa = 4.13DD51 pKa = 4.31KK52 pKa = 11.46YY53 pKa = 11.4DD54 pKa = 3.36YY55 pKa = 11.47KK56 pKa = 11.17LIEE59 pKa = 5.22DD60 pKa = 4.71KK61 pKa = 11.07LSKK64 pKa = 10.98CNFDD68 pKa = 3.66YY69 pKa = 11.2EE70 pKa = 4.41IVSFFTYY77 pKa = 10.63SDD79 pKa = 3.12QKK81 pKa = 10.83YY82 pKa = 11.14DD83 pKa = 3.6NVEE86 pKa = 3.94QKK88 pKa = 10.68LLKK91 pKa = 10.26YY92 pKa = 10.1VNKK95 pKa = 10.41KK96 pKa = 9.74KK97 pKa = 11.02VLVADD102 pKa = 4.05LTRR105 pKa = 11.84NKK107 pKa = 10.33LKK109 pKa = 11.05YY110 pKa = 8.77EE111 pKa = 3.95NNYY114 pKa = 7.37ITKK117 pKa = 10.34DD118 pKa = 3.29LYY120 pKa = 11.08TEE122 pKa = 4.97DD123 pKa = 5.74DD124 pKa = 3.94YY125 pKa = 11.89TDD127 pKa = 3.45TLMDD131 pKa = 4.3PSINTSLRR139 pKa = 11.84SNLNAAMFVIEE150 pKa = 4.73NIEE153 pKa = 4.03DD154 pKa = 3.44KK155 pKa = 10.71KK156 pKa = 9.84IQSRR160 pKa = 11.84YY161 pKa = 7.2RR162 pKa = 11.84HH163 pKa = 5.57LFEE166 pKa = 4.84YY167 pKa = 10.29VASTVSHH174 pKa = 5.99YY175 pKa = 10.49GIPVHH180 pKa = 5.21NQKK183 pKa = 9.88YY184 pKa = 8.84RR185 pKa = 11.84YY186 pKa = 9.29DD187 pKa = 3.71YY188 pKa = 11.44KK189 pKa = 11.35EE190 pKa = 4.35MYY192 pKa = 9.24NKK194 pKa = 10.14PYY196 pKa = 11.02YY197 pKa = 9.89LVPWANSSIEE207 pKa = 4.05MIFAVSSHH215 pKa = 5.47EE216 pKa = 4.15LYY218 pKa = 10.87LVAEE222 pKa = 4.21EE223 pKa = 5.11LIINSFSNRR232 pKa = 11.84STLAKK237 pKa = 9.32LTSSPMTTLTSKK249 pKa = 11.12LDD251 pKa = 3.7LYY253 pKa = 10.29SCYY256 pKa = 8.38MTTEE260 pKa = 4.21DD261 pKa = 6.13LEE263 pKa = 5.03LEE265 pKa = 4.24YY266 pKa = 10.84SDD268 pKa = 5.93KK269 pKa = 10.73IVKK272 pKa = 10.24SYY274 pKa = 9.05ITDD277 pKa = 3.53EE278 pKa = 4.36AQLEE282 pKa = 4.23LDD284 pKa = 3.7EE285 pKa = 5.51QIAEE289 pKa = 4.06LRR291 pKa = 11.84KK292 pKa = 10.28EE293 pKa = 4.22KK294 pKa = 8.37MTKK297 pKa = 8.47TADD300 pKa = 5.39IIDD303 pKa = 3.45EE304 pKa = 4.23WQRR307 pKa = 11.84NPSIDD312 pKa = 3.93TFPLMAQIYY321 pKa = 9.28SFSFHH326 pKa = 5.86VGYY329 pKa = 9.99RR330 pKa = 11.84KK331 pKa = 10.3QMMLDD336 pKa = 3.21AVTDD340 pKa = 3.74QLSIEE345 pKa = 4.23YY346 pKa = 10.38SDD348 pKa = 4.42TFDD351 pKa = 4.1KK352 pKa = 11.3DD353 pKa = 3.37MYY355 pKa = 11.77NMYY358 pKa = 9.54TNNIIKK364 pKa = 10.23IIDD367 pKa = 3.76DD368 pKa = 3.67MLRR371 pKa = 11.84DD372 pKa = 4.17HH373 pKa = 7.15IKK375 pKa = 10.79KK376 pKa = 9.31EE377 pKa = 4.37DD378 pKa = 3.55KK379 pKa = 10.18MLLNAEE385 pKa = 4.15IAGLLSMSSASNGEE399 pKa = 3.78SRR401 pKa = 11.84VIKK404 pKa = 10.36FGRR407 pKa = 11.84RR408 pKa = 11.84TVRR411 pKa = 11.84STKK414 pKa = 10.83KK415 pKa = 10.29NMHH418 pKa = 5.59VMDD421 pKa = 6.45DD422 pKa = 4.08IFNKK426 pKa = 10.18QYY428 pKa = 11.29DD429 pKa = 4.25LDD431 pKa = 4.18VPDD434 pKa = 4.1VNEE437 pKa = 4.4NNPIPLGRR445 pKa = 11.84RR446 pKa = 11.84DD447 pKa = 3.43VPGRR451 pKa = 11.84RR452 pKa = 11.84TRR454 pKa = 11.84VIFILPYY461 pKa = 9.41PYY463 pKa = 10.35FIAQHH468 pKa = 5.56SVVEE472 pKa = 4.38LYY474 pKa = 10.78LRR476 pKa = 11.84EE477 pKa = 4.2SKK479 pKa = 10.48HH480 pKa = 6.13IKK482 pKa = 9.82EE483 pKa = 4.0FSEE486 pKa = 5.59FYY488 pKa = 10.47SQSSQLLSYY497 pKa = 11.28GDD499 pKa = 3.43TNRR502 pKa = 11.84YY503 pKa = 9.11LNQSTIIVYY512 pKa = 10.72ADD514 pKa = 3.11VSQWDD519 pKa = 3.89SSKK522 pKa = 11.38HH523 pKa = 3.63NTTPLRR529 pKa = 11.84NAILKK534 pKa = 10.42AIEE537 pKa = 3.94QLKK540 pKa = 10.77SYY542 pKa = 7.08TTNEE546 pKa = 3.89KK547 pKa = 10.91VITALNNYY555 pKa = 9.97ALTQMKK561 pKa = 9.97LKK563 pKa = 10.79NSFVTIGNKK572 pKa = 8.58VLQYY576 pKa = 10.7GAVASGEE583 pKa = 4.2KK584 pKa = 7.8QTKK587 pKa = 9.65LMNSIANLALITTVINYY604 pKa = 9.8LNVGEE609 pKa = 4.57SYY611 pKa = 10.27RR612 pKa = 11.84IKK614 pKa = 10.41IIRR617 pKa = 11.84VDD619 pKa = 3.23GDD621 pKa = 3.57DD622 pKa = 4.13NYY624 pKa = 11.28FIMEE628 pKa = 4.75FDD630 pKa = 3.67RR631 pKa = 11.84KK632 pKa = 10.01VDD634 pKa = 3.53KK635 pKa = 11.18QLVTVVSNAVKK646 pKa = 9.42DD647 pKa = 3.53TYY649 pKa = 11.66GRR651 pKa = 11.84MDD653 pKa = 3.71VKK655 pKa = 11.12VKK657 pKa = 10.83ALTATTGLEE666 pKa = 3.7MAKK669 pKa = 10.03RR670 pKa = 11.84YY671 pKa = 9.19IAGGRR676 pKa = 11.84LFFRR680 pKa = 11.84AGVNILNNEE689 pKa = 3.97KK690 pKa = 9.28RR691 pKa = 11.84THH693 pKa = 4.91TTTYY697 pKa = 9.86DD698 pKa = 3.08QAAIIYY704 pKa = 10.39ANYY707 pKa = 9.82LVNKK711 pKa = 8.05MRR713 pKa = 11.84GYY715 pKa = 9.58NMEE718 pKa = 5.28RR719 pKa = 11.84IFILCKK725 pKa = 9.57IMQMTSVKK733 pKa = 8.62ITGSIRR739 pKa = 11.84LFPASLMLTTNSPYY753 pKa = 10.77KK754 pKa = 10.93VFDD757 pKa = 4.42DD758 pKa = 3.6VDD760 pKa = 4.4FVQTYY765 pKa = 9.8NSSTIAIHH773 pKa = 5.12LQKK776 pKa = 10.22MLISVSQVKK785 pKa = 10.7SNLADD790 pKa = 5.48DD791 pKa = 3.8IAKK794 pKa = 10.13SEE796 pKa = 4.05KK797 pKa = 8.34FTNYY801 pKa = 9.3VQFLTKK807 pKa = 10.39KK808 pKa = 10.68LLVHH812 pKa = 6.14NNKK815 pKa = 9.29IVEE818 pKa = 4.07MGIARR823 pKa = 11.84TEE825 pKa = 3.97KK826 pKa = 10.71AKK828 pKa = 10.93LNSYY832 pKa = 9.89PPIANEE838 pKa = 4.31KK839 pKa = 10.45RR840 pKa = 11.84NNQLKK845 pKa = 8.53TLMTFLQTPTYY856 pKa = 10.82KK857 pKa = 10.04MSTDD861 pKa = 3.27VTINDD866 pKa = 3.16MMKK869 pKa = 10.22IINEE873 pKa = 4.09YY874 pKa = 9.51TNYY877 pKa = 10.51TIITDD882 pKa = 3.59KK883 pKa = 11.41YY884 pKa = 9.48GIQPNPMPLLPEE896 pKa = 4.82NIQFAMSHH904 pKa = 4.95VGARR908 pKa = 11.84VYY910 pKa = 10.6QIEE913 pKa = 4.06EE914 pKa = 3.9SAARR918 pKa = 11.84SAISKK923 pKa = 9.84LISNYY928 pKa = 7.6TVYY931 pKa = 10.58KK932 pKa = 10.04PSVDD936 pKa = 3.37EE937 pKa = 4.38LYY939 pKa = 10.92KK940 pKa = 10.81VINSNEE946 pKa = 4.18RR947 pKa = 11.84ILFEE951 pKa = 4.25YY952 pKa = 8.62VTSFGVPTRR961 pKa = 11.84DD962 pKa = 2.92VTAYY966 pKa = 10.72LSAKK970 pKa = 9.98LYY972 pKa = 10.89KK973 pKa = 9.66KK974 pKa = 10.35DD975 pKa = 3.52RR976 pKa = 11.84NLILEE981 pKa = 4.23SYY983 pKa = 9.41IYY985 pKa = 10.65QIMSVNYY992 pKa = 9.74NAYY995 pKa = 10.29QLFNLNSEE1003 pKa = 4.85LFDD1006 pKa = 3.74KK1007 pKa = 10.85YY1008 pKa = 11.21INIVTYY1014 pKa = 9.49MKK1016 pKa = 10.18IPSVNFIIFTYY1027 pKa = 10.3LKK1029 pKa = 10.75LVILNKK1035 pKa = 9.5MLQEE1039 pKa = 4.09RR1040 pKa = 11.84KK1041 pKa = 9.62LYY1043 pKa = 10.19KK1044 pKa = 10.15AYY1046 pKa = 10.6CQIPKK1051 pKa = 10.03HH1052 pKa = 5.83LLHH1055 pKa = 6.89ILWKK1059 pKa = 9.92MSINLATISSPYY1071 pKa = 9.2TIANFFQEE1079 pKa = 4.12
MM1 pKa = 7.01GTYY4 pKa = 10.78NLLLQKK10 pKa = 9.6YY11 pKa = 8.78LRR13 pKa = 11.84WLYY16 pKa = 11.11GKK18 pKa = 9.58DD19 pKa = 2.94ATVIPIFITNDD30 pKa = 3.14QNNQEE35 pKa = 4.26ILNSIVDD42 pKa = 3.79YY43 pKa = 11.04AKK45 pKa = 10.61QLAIDD50 pKa = 4.13DD51 pKa = 4.31KK52 pKa = 11.46YY53 pKa = 11.4DD54 pKa = 3.36YY55 pKa = 11.47KK56 pKa = 11.17LIEE59 pKa = 5.22DD60 pKa = 4.71KK61 pKa = 11.07LSKK64 pKa = 10.98CNFDD68 pKa = 3.66YY69 pKa = 11.2EE70 pKa = 4.41IVSFFTYY77 pKa = 10.63SDD79 pKa = 3.12QKK81 pKa = 10.83YY82 pKa = 11.14DD83 pKa = 3.6NVEE86 pKa = 3.94QKK88 pKa = 10.68LLKK91 pKa = 10.26YY92 pKa = 10.1VNKK95 pKa = 10.41KK96 pKa = 9.74KK97 pKa = 11.02VLVADD102 pKa = 4.05LTRR105 pKa = 11.84NKK107 pKa = 10.33LKK109 pKa = 11.05YY110 pKa = 8.77EE111 pKa = 3.95NNYY114 pKa = 7.37ITKK117 pKa = 10.34DD118 pKa = 3.29LYY120 pKa = 11.08TEE122 pKa = 4.97DD123 pKa = 5.74DD124 pKa = 3.94YY125 pKa = 11.89TDD127 pKa = 3.45TLMDD131 pKa = 4.3PSINTSLRR139 pKa = 11.84SNLNAAMFVIEE150 pKa = 4.73NIEE153 pKa = 4.03DD154 pKa = 3.44KK155 pKa = 10.71KK156 pKa = 9.84IQSRR160 pKa = 11.84YY161 pKa = 7.2RR162 pKa = 11.84HH163 pKa = 5.57LFEE166 pKa = 4.84YY167 pKa = 10.29VASTVSHH174 pKa = 5.99YY175 pKa = 10.49GIPVHH180 pKa = 5.21NQKK183 pKa = 9.88YY184 pKa = 8.84RR185 pKa = 11.84YY186 pKa = 9.29DD187 pKa = 3.71YY188 pKa = 11.44KK189 pKa = 11.35EE190 pKa = 4.35MYY192 pKa = 9.24NKK194 pKa = 10.14PYY196 pKa = 11.02YY197 pKa = 9.89LVPWANSSIEE207 pKa = 4.05MIFAVSSHH215 pKa = 5.47EE216 pKa = 4.15LYY218 pKa = 10.87LVAEE222 pKa = 4.21EE223 pKa = 5.11LIINSFSNRR232 pKa = 11.84STLAKK237 pKa = 9.32LTSSPMTTLTSKK249 pKa = 11.12LDD251 pKa = 3.7LYY253 pKa = 10.29SCYY256 pKa = 8.38MTTEE260 pKa = 4.21DD261 pKa = 6.13LEE263 pKa = 5.03LEE265 pKa = 4.24YY266 pKa = 10.84SDD268 pKa = 5.93KK269 pKa = 10.73IVKK272 pKa = 10.24SYY274 pKa = 9.05ITDD277 pKa = 3.53EE278 pKa = 4.36AQLEE282 pKa = 4.23LDD284 pKa = 3.7EE285 pKa = 5.51QIAEE289 pKa = 4.06LRR291 pKa = 11.84KK292 pKa = 10.28EE293 pKa = 4.22KK294 pKa = 8.37MTKK297 pKa = 8.47TADD300 pKa = 5.39IIDD303 pKa = 3.45EE304 pKa = 4.23WQRR307 pKa = 11.84NPSIDD312 pKa = 3.93TFPLMAQIYY321 pKa = 9.28SFSFHH326 pKa = 5.86VGYY329 pKa = 9.99RR330 pKa = 11.84KK331 pKa = 10.3QMMLDD336 pKa = 3.21AVTDD340 pKa = 3.74QLSIEE345 pKa = 4.23YY346 pKa = 10.38SDD348 pKa = 4.42TFDD351 pKa = 4.1KK352 pKa = 11.3DD353 pKa = 3.37MYY355 pKa = 11.77NMYY358 pKa = 9.54TNNIIKK364 pKa = 10.23IIDD367 pKa = 3.76DD368 pKa = 3.67MLRR371 pKa = 11.84DD372 pKa = 4.17HH373 pKa = 7.15IKK375 pKa = 10.79KK376 pKa = 9.31EE377 pKa = 4.37DD378 pKa = 3.55KK379 pKa = 10.18MLLNAEE385 pKa = 4.15IAGLLSMSSASNGEE399 pKa = 3.78SRR401 pKa = 11.84VIKK404 pKa = 10.36FGRR407 pKa = 11.84RR408 pKa = 11.84TVRR411 pKa = 11.84STKK414 pKa = 10.83KK415 pKa = 10.29NMHH418 pKa = 5.59VMDD421 pKa = 6.45DD422 pKa = 4.08IFNKK426 pKa = 10.18QYY428 pKa = 11.29DD429 pKa = 4.25LDD431 pKa = 4.18VPDD434 pKa = 4.1VNEE437 pKa = 4.4NNPIPLGRR445 pKa = 11.84RR446 pKa = 11.84DD447 pKa = 3.43VPGRR451 pKa = 11.84RR452 pKa = 11.84TRR454 pKa = 11.84VIFILPYY461 pKa = 9.41PYY463 pKa = 10.35FIAQHH468 pKa = 5.56SVVEE472 pKa = 4.38LYY474 pKa = 10.78LRR476 pKa = 11.84EE477 pKa = 4.2SKK479 pKa = 10.48HH480 pKa = 6.13IKK482 pKa = 9.82EE483 pKa = 4.0FSEE486 pKa = 5.59FYY488 pKa = 10.47SQSSQLLSYY497 pKa = 11.28GDD499 pKa = 3.43TNRR502 pKa = 11.84YY503 pKa = 9.11LNQSTIIVYY512 pKa = 10.72ADD514 pKa = 3.11VSQWDD519 pKa = 3.89SSKK522 pKa = 11.38HH523 pKa = 3.63NTTPLRR529 pKa = 11.84NAILKK534 pKa = 10.42AIEE537 pKa = 3.94QLKK540 pKa = 10.77SYY542 pKa = 7.08TTNEE546 pKa = 3.89KK547 pKa = 10.91VITALNNYY555 pKa = 9.97ALTQMKK561 pKa = 9.97LKK563 pKa = 10.79NSFVTIGNKK572 pKa = 8.58VLQYY576 pKa = 10.7GAVASGEE583 pKa = 4.2KK584 pKa = 7.8QTKK587 pKa = 9.65LMNSIANLALITTVINYY604 pKa = 9.8LNVGEE609 pKa = 4.57SYY611 pKa = 10.27RR612 pKa = 11.84IKK614 pKa = 10.41IIRR617 pKa = 11.84VDD619 pKa = 3.23GDD621 pKa = 3.57DD622 pKa = 4.13NYY624 pKa = 11.28FIMEE628 pKa = 4.75FDD630 pKa = 3.67RR631 pKa = 11.84KK632 pKa = 10.01VDD634 pKa = 3.53KK635 pKa = 11.18QLVTVVSNAVKK646 pKa = 9.42DD647 pKa = 3.53TYY649 pKa = 11.66GRR651 pKa = 11.84MDD653 pKa = 3.71VKK655 pKa = 11.12VKK657 pKa = 10.83ALTATTGLEE666 pKa = 3.7MAKK669 pKa = 10.03RR670 pKa = 11.84YY671 pKa = 9.19IAGGRR676 pKa = 11.84LFFRR680 pKa = 11.84AGVNILNNEE689 pKa = 3.97KK690 pKa = 9.28RR691 pKa = 11.84THH693 pKa = 4.91TTTYY697 pKa = 9.86DD698 pKa = 3.08QAAIIYY704 pKa = 10.39ANYY707 pKa = 9.82LVNKK711 pKa = 8.05MRR713 pKa = 11.84GYY715 pKa = 9.58NMEE718 pKa = 5.28RR719 pKa = 11.84IFILCKK725 pKa = 9.57IMQMTSVKK733 pKa = 8.62ITGSIRR739 pKa = 11.84LFPASLMLTTNSPYY753 pKa = 10.77KK754 pKa = 10.93VFDD757 pKa = 4.42DD758 pKa = 3.6VDD760 pKa = 4.4FVQTYY765 pKa = 9.8NSSTIAIHH773 pKa = 5.12LQKK776 pKa = 10.22MLISVSQVKK785 pKa = 10.7SNLADD790 pKa = 5.48DD791 pKa = 3.8IAKK794 pKa = 10.13SEE796 pKa = 4.05KK797 pKa = 8.34FTNYY801 pKa = 9.3VQFLTKK807 pKa = 10.39KK808 pKa = 10.68LLVHH812 pKa = 6.14NNKK815 pKa = 9.29IVEE818 pKa = 4.07MGIARR823 pKa = 11.84TEE825 pKa = 3.97KK826 pKa = 10.71AKK828 pKa = 10.93LNSYY832 pKa = 9.89PPIANEE838 pKa = 4.31KK839 pKa = 10.45RR840 pKa = 11.84NNQLKK845 pKa = 8.53TLMTFLQTPTYY856 pKa = 10.82KK857 pKa = 10.04MSTDD861 pKa = 3.27VTINDD866 pKa = 3.16MMKK869 pKa = 10.22IINEE873 pKa = 4.09YY874 pKa = 9.51TNYY877 pKa = 10.51TIITDD882 pKa = 3.59KK883 pKa = 11.41YY884 pKa = 9.48GIQPNPMPLLPEE896 pKa = 4.82NIQFAMSHH904 pKa = 4.95VGARR908 pKa = 11.84VYY910 pKa = 10.6QIEE913 pKa = 4.06EE914 pKa = 3.9SAARR918 pKa = 11.84SAISKK923 pKa = 9.84LISNYY928 pKa = 7.6TVYY931 pKa = 10.58KK932 pKa = 10.04PSVDD936 pKa = 3.37EE937 pKa = 4.38LYY939 pKa = 10.92KK940 pKa = 10.81VINSNEE946 pKa = 4.18RR947 pKa = 11.84ILFEE951 pKa = 4.25YY952 pKa = 8.62VTSFGVPTRR961 pKa = 11.84DD962 pKa = 2.92VTAYY966 pKa = 10.72LSAKK970 pKa = 9.98LYY972 pKa = 10.89KK973 pKa = 9.66KK974 pKa = 10.35DD975 pKa = 3.52RR976 pKa = 11.84NLILEE981 pKa = 4.23SYY983 pKa = 9.41IYY985 pKa = 10.65QIMSVNYY992 pKa = 9.74NAYY995 pKa = 10.29QLFNLNSEE1003 pKa = 4.85LFDD1006 pKa = 3.74KK1007 pKa = 10.85YY1008 pKa = 11.21INIVTYY1014 pKa = 9.49MKK1016 pKa = 10.18IPSVNFIIFTYY1027 pKa = 10.3LKK1029 pKa = 10.75LVILNKK1035 pKa = 9.5MLQEE1039 pKa = 4.09RR1040 pKa = 11.84KK1041 pKa = 9.62LYY1043 pKa = 10.19KK1044 pKa = 10.15AYY1046 pKa = 10.6CQIPKK1051 pKa = 10.03HH1052 pKa = 5.83LLHH1055 pKa = 6.89ILWKK1059 pKa = 9.92MSINLATISSPYY1071 pKa = 9.2TIANFFQEE1079 pKa = 4.12
Molecular weight: 125.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5837 |
93 |
1079 |
486.4 |
56.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.985 ± 0.528 | 1.285 ± 0.432 |
6.476 ± 0.295 | 5.362 ± 0.333 |
4.095 ± 0.261 | 3.135 ± 0.332 |
1.559 ± 0.24 | 8.103 ± 0.382 |
6.39 ± 0.648 | 9.56 ± 0.247 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.964 ± 0.35 | 7.487 ± 0.335 |
2.861 ± 0.423 | 3.701 ± 0.23 |
4.934 ± 0.426 | 7.949 ± 0.432 |
6.664 ± 0.411 | 6.339 ± 0.29 |
0.685 ± 0.117 | 5.465 ± 0.447 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
