
Photobacterium marinum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Photobacterium
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
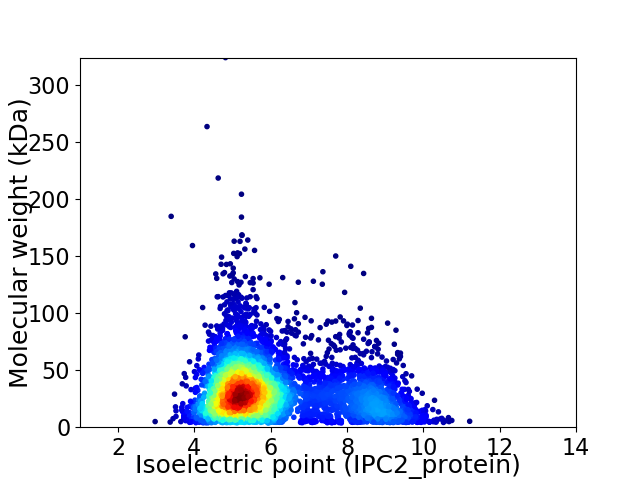
Virtual 2D-PAGE plot for 4904 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8JK54|L8JK54_9GAMM Arginine/ornithine antiporter ArcD OS=Photobacterium marinum OX=1056511 GN=C942_00187 PE=3 SV=1
MM1 pKa = 8.13AEE3 pKa = 3.52ITLPQGAPSTCKK15 pKa = 10.31AGEE18 pKa = 4.22TVSQKK23 pKa = 11.32LNFKK27 pKa = 8.64VTSTANIRR35 pKa = 11.84YY36 pKa = 8.53NFSFYY41 pKa = 8.48TTTNPLANPLDD52 pKa = 4.16GVPGGNSNEE61 pKa = 3.88CLIWVGEE68 pKa = 4.12VGDD71 pKa = 5.18AGGLPNSQSANGDD84 pKa = 3.22LCADD88 pKa = 3.56VTKK91 pKa = 10.94SQDD94 pKa = 2.66AVYY97 pKa = 9.83QEE99 pKa = 4.13EE100 pKa = 5.11VITFFCQDD108 pKa = 2.75TDD110 pKa = 3.34GDD112 pKa = 4.37SFVDD116 pKa = 4.54LDD118 pKa = 4.6YY119 pKa = 11.48CATWDD124 pKa = 3.83QNDD127 pKa = 4.5DD128 pKa = 3.99NLCSSGGEE136 pKa = 4.07IQSGLLPAPGAPSKK150 pKa = 10.76CNCEE154 pKa = 3.99TVTIPVKK161 pKa = 10.5VLPSEE166 pKa = 4.19PVLEE170 pKa = 4.14KK171 pKa = 10.69AINGDD176 pKa = 3.24ATRR179 pKa = 11.84PEE181 pKa = 3.93SDD183 pKa = 2.77IFAGNDD189 pKa = 3.22SFSFDD194 pKa = 4.33LKK196 pKa = 9.52VTNPNTNTSLVITEE210 pKa = 4.29LVEE213 pKa = 4.09KK214 pKa = 10.76FGGEE218 pKa = 3.84EE219 pKa = 3.92FLISGSLSSSTVSLNEE235 pKa = 3.97DD236 pKa = 3.07QVVLVNAVDD245 pKa = 3.45NDD247 pKa = 3.73GRR249 pKa = 11.84CLASGSEE256 pKa = 4.34TIAPGATYY264 pKa = 10.1ICTVTLRR271 pKa = 11.84WKK273 pKa = 10.77SLILVDD279 pKa = 3.89TDD281 pKa = 3.82SSIEE285 pKa = 3.91GVRR288 pKa = 11.84EE289 pKa = 3.93DD290 pKa = 3.85KK291 pKa = 11.03VSQFGVHH298 pKa = 5.52WKK300 pKa = 10.42FDD302 pKa = 4.4DD303 pKa = 4.59GPEE306 pKa = 4.1AYY308 pKa = 8.91MGPSNSLTVSISDD321 pKa = 3.62VPPIISIAKK330 pKa = 8.04VASPTSIPEE339 pKa = 3.91TGPTGFDD346 pKa = 3.24YY347 pKa = 10.78VHH349 pKa = 6.81YY350 pKa = 10.0YY351 pKa = 8.11VTYY354 pKa = 9.96SNEE357 pKa = 4.23SGWDD361 pKa = 3.48QIHH364 pKa = 6.6VKK366 pKa = 9.47DD367 pKa = 3.92TDD369 pKa = 4.05FIDD372 pKa = 5.12LVQEE376 pKa = 4.09MNTISEE382 pKa = 4.53PKK384 pKa = 8.31PVGYY388 pKa = 8.54TGCAINGLTYY398 pKa = 9.78TQGSGCDD405 pKa = 3.3YY406 pKa = 9.88GVNMGTLYY414 pKa = 11.09SSINAGDD421 pKa = 3.78VYY423 pKa = 11.3KK424 pKa = 10.19NTVKK428 pKa = 10.79VIPTDD433 pKa = 3.44EE434 pKa = 4.57EE435 pKa = 4.65GTSGEE440 pKa = 4.73LKK442 pKa = 9.51TAAASISVEE451 pKa = 3.86NVNPVVTLKK460 pKa = 10.72KK461 pKa = 10.17YY462 pKa = 10.27VRR464 pKa = 11.84AGLAPEE470 pKa = 4.36VNPLDD475 pKa = 3.71PAGYY479 pKa = 9.68HH480 pKa = 7.0DD481 pKa = 5.01MSTSVDD487 pKa = 3.62EE488 pKa = 4.47YY489 pKa = 11.43QLTDD493 pKa = 3.81LSRR496 pKa = 11.84APVVTFLFVVSNSSFEE512 pKa = 4.03NFEE515 pKa = 3.73IMDD518 pKa = 3.96FVDD521 pKa = 4.84FAQAEE526 pKa = 4.72TFGLNPPAQVFDD538 pKa = 4.36SNIADD543 pKa = 3.95TVPVDD548 pKa = 3.98DD549 pKa = 4.7TCSGLIGSSVAVNEE563 pKa = 4.47SVACTMSFKK572 pKa = 10.94VAGDD576 pKa = 4.09DD577 pKa = 4.05SDD579 pKa = 4.82SVDD582 pKa = 3.45NIAWVLVSDD591 pKa = 4.94GEE593 pKa = 4.32QEE595 pKa = 3.98AGGSIGQGFDD605 pKa = 2.93TDD607 pKa = 3.52DD608 pKa = 4.06AMVEE612 pKa = 4.07FNPVGNSIALGLDD625 pKa = 3.19MSATVKK631 pKa = 10.83LSVTAHH637 pKa = 6.6ADD639 pKa = 3.37NVEE642 pKa = 4.14VLSFEE647 pKa = 4.31PLSDD651 pKa = 4.0VIVMVATEE659 pKa = 4.7DD660 pKa = 3.64PNAAEE665 pKa = 4.34TTLDD669 pKa = 3.51EE670 pKa = 4.77PLFSGTFDD678 pKa = 3.0QFTVANIDD686 pKa = 3.77CSSTTIDD693 pKa = 3.03PGYY696 pKa = 9.43TYY698 pKa = 10.86SCSFTFTPKK707 pKa = 10.89GEE709 pKa = 4.2YY710 pKa = 8.61TATAGLQVLNDD721 pKa = 3.8TLKK724 pKa = 10.72VRR726 pKa = 11.84VRR728 pKa = 11.84DD729 pKa = 3.61DD730 pKa = 4.01DD731 pKa = 3.96GTVQEE736 pKa = 4.76LSAVITVEE744 pKa = 3.97AQQ746 pKa = 2.76
MM1 pKa = 8.13AEE3 pKa = 3.52ITLPQGAPSTCKK15 pKa = 10.31AGEE18 pKa = 4.22TVSQKK23 pKa = 11.32LNFKK27 pKa = 8.64VTSTANIRR35 pKa = 11.84YY36 pKa = 8.53NFSFYY41 pKa = 8.48TTTNPLANPLDD52 pKa = 4.16GVPGGNSNEE61 pKa = 3.88CLIWVGEE68 pKa = 4.12VGDD71 pKa = 5.18AGGLPNSQSANGDD84 pKa = 3.22LCADD88 pKa = 3.56VTKK91 pKa = 10.94SQDD94 pKa = 2.66AVYY97 pKa = 9.83QEE99 pKa = 4.13EE100 pKa = 5.11VITFFCQDD108 pKa = 2.75TDD110 pKa = 3.34GDD112 pKa = 4.37SFVDD116 pKa = 4.54LDD118 pKa = 4.6YY119 pKa = 11.48CATWDD124 pKa = 3.83QNDD127 pKa = 4.5DD128 pKa = 3.99NLCSSGGEE136 pKa = 4.07IQSGLLPAPGAPSKK150 pKa = 10.76CNCEE154 pKa = 3.99TVTIPVKK161 pKa = 10.5VLPSEE166 pKa = 4.19PVLEE170 pKa = 4.14KK171 pKa = 10.69AINGDD176 pKa = 3.24ATRR179 pKa = 11.84PEE181 pKa = 3.93SDD183 pKa = 2.77IFAGNDD189 pKa = 3.22SFSFDD194 pKa = 4.33LKK196 pKa = 9.52VTNPNTNTSLVITEE210 pKa = 4.29LVEE213 pKa = 4.09KK214 pKa = 10.76FGGEE218 pKa = 3.84EE219 pKa = 3.92FLISGSLSSSTVSLNEE235 pKa = 3.97DD236 pKa = 3.07QVVLVNAVDD245 pKa = 3.45NDD247 pKa = 3.73GRR249 pKa = 11.84CLASGSEE256 pKa = 4.34TIAPGATYY264 pKa = 10.1ICTVTLRR271 pKa = 11.84WKK273 pKa = 10.77SLILVDD279 pKa = 3.89TDD281 pKa = 3.82SSIEE285 pKa = 3.91GVRR288 pKa = 11.84EE289 pKa = 3.93DD290 pKa = 3.85KK291 pKa = 11.03VSQFGVHH298 pKa = 5.52WKK300 pKa = 10.42FDD302 pKa = 4.4DD303 pKa = 4.59GPEE306 pKa = 4.1AYY308 pKa = 8.91MGPSNSLTVSISDD321 pKa = 3.62VPPIISIAKK330 pKa = 8.04VASPTSIPEE339 pKa = 3.91TGPTGFDD346 pKa = 3.24YY347 pKa = 10.78VHH349 pKa = 6.81YY350 pKa = 10.0YY351 pKa = 8.11VTYY354 pKa = 9.96SNEE357 pKa = 4.23SGWDD361 pKa = 3.48QIHH364 pKa = 6.6VKK366 pKa = 9.47DD367 pKa = 3.92TDD369 pKa = 4.05FIDD372 pKa = 5.12LVQEE376 pKa = 4.09MNTISEE382 pKa = 4.53PKK384 pKa = 8.31PVGYY388 pKa = 8.54TGCAINGLTYY398 pKa = 9.78TQGSGCDD405 pKa = 3.3YY406 pKa = 9.88GVNMGTLYY414 pKa = 11.09SSINAGDD421 pKa = 3.78VYY423 pKa = 11.3KK424 pKa = 10.19NTVKK428 pKa = 10.79VIPTDD433 pKa = 3.44EE434 pKa = 4.57EE435 pKa = 4.65GTSGEE440 pKa = 4.73LKK442 pKa = 9.51TAAASISVEE451 pKa = 3.86NVNPVVTLKK460 pKa = 10.72KK461 pKa = 10.17YY462 pKa = 10.27VRR464 pKa = 11.84AGLAPEE470 pKa = 4.36VNPLDD475 pKa = 3.71PAGYY479 pKa = 9.68HH480 pKa = 7.0DD481 pKa = 5.01MSTSVDD487 pKa = 3.62EE488 pKa = 4.47YY489 pKa = 11.43QLTDD493 pKa = 3.81LSRR496 pKa = 11.84APVVTFLFVVSNSSFEE512 pKa = 4.03NFEE515 pKa = 3.73IMDD518 pKa = 3.96FVDD521 pKa = 4.84FAQAEE526 pKa = 4.72TFGLNPPAQVFDD538 pKa = 4.36SNIADD543 pKa = 3.95TVPVDD548 pKa = 3.98DD549 pKa = 4.7TCSGLIGSSVAVNEE563 pKa = 4.47SVACTMSFKK572 pKa = 10.94VAGDD576 pKa = 4.09DD577 pKa = 4.05SDD579 pKa = 4.82SVDD582 pKa = 3.45NIAWVLVSDD591 pKa = 4.94GEE593 pKa = 4.32QEE595 pKa = 3.98AGGSIGQGFDD605 pKa = 2.93TDD607 pKa = 3.52DD608 pKa = 4.06AMVEE612 pKa = 4.07FNPVGNSIALGLDD625 pKa = 3.19MSATVKK631 pKa = 10.83LSVTAHH637 pKa = 6.6ADD639 pKa = 3.37NVEE642 pKa = 4.14VLSFEE647 pKa = 4.31PLSDD651 pKa = 4.0VIVMVATEE659 pKa = 4.7DD660 pKa = 3.64PNAAEE665 pKa = 4.34TTLDD669 pKa = 3.51EE670 pKa = 4.77PLFSGTFDD678 pKa = 3.0QFTVANIDD686 pKa = 3.77CSSTTIDD693 pKa = 3.03PGYY696 pKa = 9.43TYY698 pKa = 10.86SCSFTFTPKK707 pKa = 10.89GEE709 pKa = 4.2YY710 pKa = 8.61TATAGLQVLNDD721 pKa = 3.8TLKK724 pKa = 10.72VRR726 pKa = 11.84VRR728 pKa = 11.84DD729 pKa = 3.61DD730 pKa = 4.01DD731 pKa = 3.96GTVQEE736 pKa = 4.76LSAVITVEE744 pKa = 3.97AQQ746 pKa = 2.76
Molecular weight: 79.23 kDa
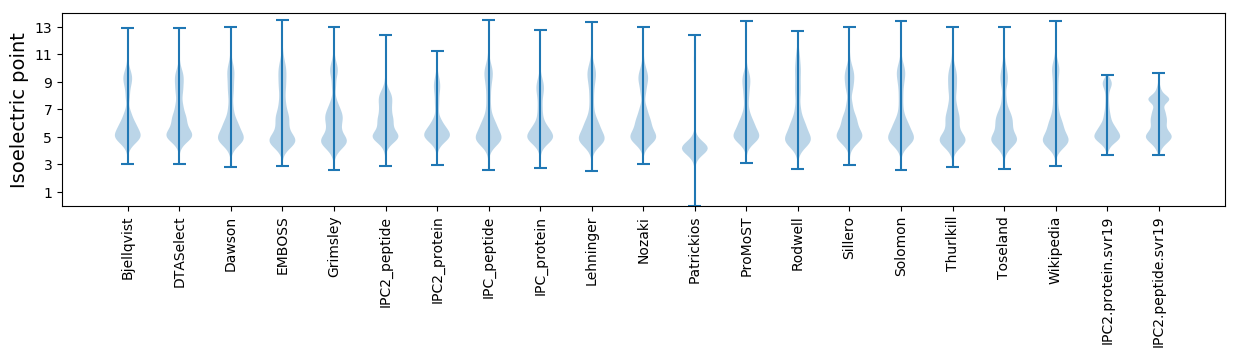
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8J655|L8J655_9GAMM Uncharacterized protein OS=Photobacterium marinum OX=1056511 GN=C942_02582 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84ATINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44NGRR28 pKa = 11.84ATINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSKK44 pKa = 10.84
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1526020 |
37 |
2918 |
311.2 |
34.6 |
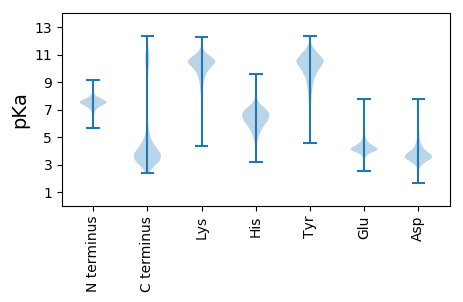
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.7 ± 0.038 | 1.129 ± 0.016 |
5.533 ± 0.03 | 6.518 ± 0.036 |
4.029 ± 0.031 | 6.863 ± 0.032 |
2.236 ± 0.019 | 6.23 ± 0.029 |
5.266 ± 0.03 | 10.335 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.775 ± 0.02 | 4.227 ± 0.025 |
3.984 ± 0.022 | 4.507 ± 0.03 |
4.745 ± 0.03 | 6.432 ± 0.03 |
5.256 ± 0.024 | 6.943 ± 0.031 |
1.247 ± 0.015 | 3.045 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
