
Cronartium ribicola mitovirus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.68
Get precalculated fractions of proteins
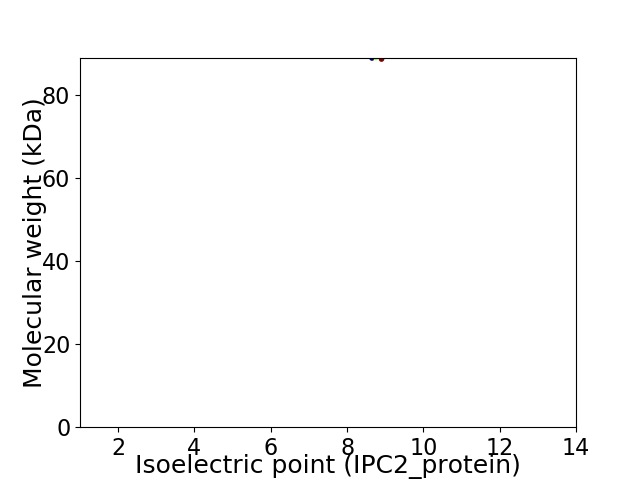
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A191KCP0|A0A191KCP0_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 5 OX=1816488 GN=RdRp PE=4 SV=1
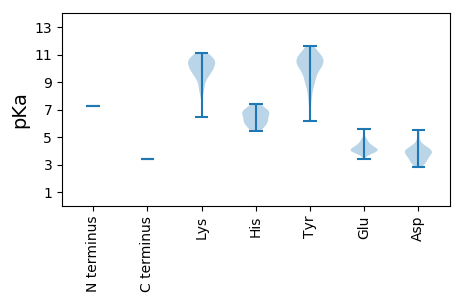
MM1 pKa = 7.27SRR3 pKa = 11.84KK4 pKa = 8.92EE5 pKa = 3.97LYY7 pKa = 10.32LVNQSFKK14 pKa = 11.08SINNCSIIDD23 pKa = 3.83KK24 pKa = 10.58LKK26 pKa = 10.68LGPQVRR32 pKa = 11.84ILIWLAVKK40 pKa = 10.63VGLKK44 pKa = 10.11PSLSIQYY51 pKa = 7.94ITWFVTLTEE60 pKa = 4.06QVGIKK65 pKa = 9.42QAIQINKK72 pKa = 9.63SIRR75 pKa = 11.84LAFTRR80 pKa = 11.84WVCGDD85 pKa = 3.1TTIRR89 pKa = 11.84IEE91 pKa = 4.04KK92 pKa = 8.54VAVYY96 pKa = 10.61KK97 pKa = 10.78DD98 pKa = 4.19GYY100 pKa = 9.11PKK102 pKa = 10.56RR103 pKa = 11.84LRR105 pKa = 11.84QFRR108 pKa = 11.84NLIKK112 pKa = 9.92EE113 pKa = 4.17DD114 pKa = 4.16PSIKK118 pKa = 10.47SYY120 pKa = 11.09ILTLLNITRR129 pKa = 11.84SLEE132 pKa = 4.03LPLTLEE138 pKa = 4.42LSSVQDD144 pKa = 3.9PFTGLQTNIEE154 pKa = 4.4DD155 pKa = 3.97MYY157 pKa = 11.63KK158 pKa = 10.57DD159 pKa = 2.99IPKK162 pKa = 9.47FLNYY166 pKa = 9.23IRR168 pKa = 11.84SRR170 pKa = 11.84PGRR173 pKa = 11.84GKK175 pKa = 9.72IEE177 pKa = 4.07FKK179 pKa = 10.2KK180 pKa = 9.64WHH182 pKa = 6.01FTTKK186 pKa = 10.4NGPNGHH192 pKa = 7.4AYY194 pKa = 6.16TTMYY198 pKa = 10.79EE199 pKa = 4.09EE200 pKa = 5.56LRR202 pKa = 11.84TCSEE206 pKa = 4.54EE207 pKa = 3.94IIEE210 pKa = 4.71SISIVGGPKK219 pKa = 9.47IKK221 pKa = 10.61EE222 pKa = 4.07KK223 pKa = 10.84INDD226 pKa = 2.92IRR228 pKa = 11.84KK229 pKa = 9.25ILDD232 pKa = 3.8DD233 pKa = 3.65PTVYY237 pKa = 10.85SVMKK241 pKa = 10.33EE242 pKa = 3.92SLKK245 pKa = 10.71LKK247 pKa = 10.06EE248 pKa = 3.82ISEE251 pKa = 4.27PKK253 pKa = 9.05QAKK256 pKa = 8.33LTLIPDD262 pKa = 3.72KK263 pKa = 10.69EE264 pKa = 4.37GKK266 pKa = 8.08TRR268 pKa = 11.84VIGVGSYY275 pKa = 7.46WVQAGLKK282 pKa = 8.44PLHH285 pKa = 6.87DD286 pKa = 4.08FVMTVIRR293 pKa = 11.84RR294 pKa = 11.84IPGDD298 pKa = 3.19CTYY301 pKa = 10.47FQEE304 pKa = 4.53RR305 pKa = 11.84APKK308 pKa = 9.85ILRR311 pKa = 11.84KK312 pKa = 9.72QPGHH316 pKa = 6.59HH317 pKa = 6.06YY318 pKa = 10.2WSFDD322 pKa = 3.29LSSATDD328 pKa = 3.59RR329 pKa = 11.84FPRR332 pKa = 11.84VLQSKK337 pKa = 10.0IIASIYY343 pKa = 8.84NDD345 pKa = 3.89KK346 pKa = 10.78IATGWLTLISLPFHH360 pKa = 6.87FRR362 pKa = 11.84GQEE365 pKa = 3.41VKK367 pKa = 10.71YY368 pKa = 10.82AVGQPIGFYY377 pKa = 10.58SSWPVFTLTHH387 pKa = 6.85HH388 pKa = 5.6YY389 pKa = 9.52CVWVACKK396 pKa = 10.06RR397 pKa = 11.84AGVSPKK403 pKa = 10.5GVYY406 pKa = 10.25ALLGDD411 pKa = 5.48DD412 pKa = 5.13IVICHH417 pKa = 6.84DD418 pKa = 4.29LVAVKK423 pKa = 10.3YY424 pKa = 10.71LEE426 pKa = 4.91IISDD430 pKa = 4.13LGVDD434 pKa = 3.35ISKK437 pKa = 10.72QKK439 pKa = 6.43THH441 pKa = 6.95KK442 pKa = 10.79SSTCYY447 pKa = 10.19EE448 pKa = 4.18FAKK451 pKa = 10.12RR452 pKa = 11.84WYY454 pKa = 10.13QEE456 pKa = 3.97GDD458 pKa = 4.06SEE460 pKa = 4.33WSHH463 pKa = 6.26YY464 pKa = 9.27PLSGLRR470 pKa = 11.84PNAKK474 pKa = 8.79FTDD477 pKa = 3.96VLASTLQSRR486 pKa = 11.84SRR488 pKa = 11.84GWRR491 pKa = 11.84ISDD494 pKa = 3.77LTPVEE499 pKa = 3.71QASYY503 pKa = 10.46INCLIRR509 pKa = 11.84GWSSEE514 pKa = 3.78YY515 pKa = 10.37HH516 pKa = 5.98LSYY519 pKa = 10.29SINWFTVYY527 pKa = 9.07YY528 pKa = 9.95TSYY531 pKa = 10.62CAIRR535 pKa = 11.84DD536 pKa = 3.68FTTWMSPLRR545 pKa = 11.84EE546 pKa = 4.11LWRR549 pKa = 11.84ISPKK553 pKa = 10.02IQIVNPSYY561 pKa = 10.99DD562 pKa = 3.24HH563 pKa = 6.53PQAGEE568 pKa = 3.82HH569 pKa = 6.09FYY571 pKa = 10.92EE572 pKa = 4.81GLSARR577 pKa = 11.84VFALNGLSKK586 pKa = 10.58ILTASPKK593 pKa = 9.86ARR595 pKa = 11.84KK596 pKa = 9.45LFEE599 pKa = 4.58PLVKK603 pKa = 10.18FDD605 pKa = 4.1WYY607 pKa = 10.45FDD609 pKa = 4.61PIITDD614 pKa = 4.45DD615 pKa = 3.9EE616 pKa = 4.82NEE618 pKa = 4.16AQPLILPQVEE628 pKa = 4.07VLRR631 pKa = 11.84RR632 pKa = 11.84LNTSYY637 pKa = 11.08QSARR641 pKa = 11.84WEE643 pKa = 3.93EE644 pKa = 3.85LQGFFTGKK652 pKa = 10.36GPEE655 pKa = 4.11ANLSILVCDD664 pKa = 4.1ISTLFTTRR672 pKa = 11.84NQNLSWTVLNVLVKK686 pKa = 10.59EE687 pKa = 4.38MIKK690 pKa = 10.07VMKK693 pKa = 10.06IVRR696 pKa = 11.84HH697 pKa = 5.42TGFDD701 pKa = 3.66YY702 pKa = 11.09SSEE705 pKa = 4.28EE706 pKa = 4.02PLSSQLRR713 pKa = 11.84SAIYY717 pKa = 10.08RR718 pKa = 11.84DD719 pKa = 3.36IILNVDD725 pKa = 2.85TDD727 pKa = 4.04LFINRR732 pKa = 11.84NPITKK737 pKa = 9.74YY738 pKa = 9.09YY739 pKa = 8.36YY740 pKa = 10.45ARR742 pKa = 11.84EE743 pKa = 3.84IEE745 pKa = 4.04DD746 pKa = 3.4AKK748 pKa = 11.11RR749 pKa = 11.84EE750 pKa = 4.24GVVLPSKK757 pKa = 9.44PTGSQTRR764 pKa = 11.84ILGGDD769 pKa = 3.27KK770 pKa = 10.95VSS772 pKa = 3.38
MM1 pKa = 7.27SRR3 pKa = 11.84KK4 pKa = 8.92EE5 pKa = 3.97LYY7 pKa = 10.32LVNQSFKK14 pKa = 11.08SINNCSIIDD23 pKa = 3.83KK24 pKa = 10.58LKK26 pKa = 10.68LGPQVRR32 pKa = 11.84ILIWLAVKK40 pKa = 10.63VGLKK44 pKa = 10.11PSLSIQYY51 pKa = 7.94ITWFVTLTEE60 pKa = 4.06QVGIKK65 pKa = 9.42QAIQINKK72 pKa = 9.63SIRR75 pKa = 11.84LAFTRR80 pKa = 11.84WVCGDD85 pKa = 3.1TTIRR89 pKa = 11.84IEE91 pKa = 4.04KK92 pKa = 8.54VAVYY96 pKa = 10.61KK97 pKa = 10.78DD98 pKa = 4.19GYY100 pKa = 9.11PKK102 pKa = 10.56RR103 pKa = 11.84LRR105 pKa = 11.84QFRR108 pKa = 11.84NLIKK112 pKa = 9.92EE113 pKa = 4.17DD114 pKa = 4.16PSIKK118 pKa = 10.47SYY120 pKa = 11.09ILTLLNITRR129 pKa = 11.84SLEE132 pKa = 4.03LPLTLEE138 pKa = 4.42LSSVQDD144 pKa = 3.9PFTGLQTNIEE154 pKa = 4.4DD155 pKa = 3.97MYY157 pKa = 11.63KK158 pKa = 10.57DD159 pKa = 2.99IPKK162 pKa = 9.47FLNYY166 pKa = 9.23IRR168 pKa = 11.84SRR170 pKa = 11.84PGRR173 pKa = 11.84GKK175 pKa = 9.72IEE177 pKa = 4.07FKK179 pKa = 10.2KK180 pKa = 9.64WHH182 pKa = 6.01FTTKK186 pKa = 10.4NGPNGHH192 pKa = 7.4AYY194 pKa = 6.16TTMYY198 pKa = 10.79EE199 pKa = 4.09EE200 pKa = 5.56LRR202 pKa = 11.84TCSEE206 pKa = 4.54EE207 pKa = 3.94IIEE210 pKa = 4.71SISIVGGPKK219 pKa = 9.47IKK221 pKa = 10.61EE222 pKa = 4.07KK223 pKa = 10.84INDD226 pKa = 2.92IRR228 pKa = 11.84KK229 pKa = 9.25ILDD232 pKa = 3.8DD233 pKa = 3.65PTVYY237 pKa = 10.85SVMKK241 pKa = 10.33EE242 pKa = 3.92SLKK245 pKa = 10.71LKK247 pKa = 10.06EE248 pKa = 3.82ISEE251 pKa = 4.27PKK253 pKa = 9.05QAKK256 pKa = 8.33LTLIPDD262 pKa = 3.72KK263 pKa = 10.69EE264 pKa = 4.37GKK266 pKa = 8.08TRR268 pKa = 11.84VIGVGSYY275 pKa = 7.46WVQAGLKK282 pKa = 8.44PLHH285 pKa = 6.87DD286 pKa = 4.08FVMTVIRR293 pKa = 11.84RR294 pKa = 11.84IPGDD298 pKa = 3.19CTYY301 pKa = 10.47FQEE304 pKa = 4.53RR305 pKa = 11.84APKK308 pKa = 9.85ILRR311 pKa = 11.84KK312 pKa = 9.72QPGHH316 pKa = 6.59HH317 pKa = 6.06YY318 pKa = 10.2WSFDD322 pKa = 3.29LSSATDD328 pKa = 3.59RR329 pKa = 11.84FPRR332 pKa = 11.84VLQSKK337 pKa = 10.0IIASIYY343 pKa = 8.84NDD345 pKa = 3.89KK346 pKa = 10.78IATGWLTLISLPFHH360 pKa = 6.87FRR362 pKa = 11.84GQEE365 pKa = 3.41VKK367 pKa = 10.71YY368 pKa = 10.82AVGQPIGFYY377 pKa = 10.58SSWPVFTLTHH387 pKa = 6.85HH388 pKa = 5.6YY389 pKa = 9.52CVWVACKK396 pKa = 10.06RR397 pKa = 11.84AGVSPKK403 pKa = 10.5GVYY406 pKa = 10.25ALLGDD411 pKa = 5.48DD412 pKa = 5.13IVICHH417 pKa = 6.84DD418 pKa = 4.29LVAVKK423 pKa = 10.3YY424 pKa = 10.71LEE426 pKa = 4.91IISDD430 pKa = 4.13LGVDD434 pKa = 3.35ISKK437 pKa = 10.72QKK439 pKa = 6.43THH441 pKa = 6.95KK442 pKa = 10.79SSTCYY447 pKa = 10.19EE448 pKa = 4.18FAKK451 pKa = 10.12RR452 pKa = 11.84WYY454 pKa = 10.13QEE456 pKa = 3.97GDD458 pKa = 4.06SEE460 pKa = 4.33WSHH463 pKa = 6.26YY464 pKa = 9.27PLSGLRR470 pKa = 11.84PNAKK474 pKa = 8.79FTDD477 pKa = 3.96VLASTLQSRR486 pKa = 11.84SRR488 pKa = 11.84GWRR491 pKa = 11.84ISDD494 pKa = 3.77LTPVEE499 pKa = 3.71QASYY503 pKa = 10.46INCLIRR509 pKa = 11.84GWSSEE514 pKa = 3.78YY515 pKa = 10.37HH516 pKa = 5.98LSYY519 pKa = 10.29SINWFTVYY527 pKa = 9.07YY528 pKa = 9.95TSYY531 pKa = 10.62CAIRR535 pKa = 11.84DD536 pKa = 3.68FTTWMSPLRR545 pKa = 11.84EE546 pKa = 4.11LWRR549 pKa = 11.84ISPKK553 pKa = 10.02IQIVNPSYY561 pKa = 10.99DD562 pKa = 3.24HH563 pKa = 6.53PQAGEE568 pKa = 3.82HH569 pKa = 6.09FYY571 pKa = 10.92EE572 pKa = 4.81GLSARR577 pKa = 11.84VFALNGLSKK586 pKa = 10.58ILTASPKK593 pKa = 9.86ARR595 pKa = 11.84KK596 pKa = 9.45LFEE599 pKa = 4.58PLVKK603 pKa = 10.18FDD605 pKa = 4.1WYY607 pKa = 10.45FDD609 pKa = 4.61PIITDD614 pKa = 4.45DD615 pKa = 3.9EE616 pKa = 4.82NEE618 pKa = 4.16AQPLILPQVEE628 pKa = 4.07VLRR631 pKa = 11.84RR632 pKa = 11.84LNTSYY637 pKa = 11.08QSARR641 pKa = 11.84WEE643 pKa = 3.93EE644 pKa = 3.85LQGFFTGKK652 pKa = 10.36GPEE655 pKa = 4.11ANLSILVCDD664 pKa = 4.1ISTLFTTRR672 pKa = 11.84NQNLSWTVLNVLVKK686 pKa = 10.59EE687 pKa = 4.38MIKK690 pKa = 10.07VMKK693 pKa = 10.06IVRR696 pKa = 11.84HH697 pKa = 5.42TGFDD701 pKa = 3.66YY702 pKa = 11.09SSEE705 pKa = 4.28EE706 pKa = 4.02PLSSQLRR713 pKa = 11.84SAIYY717 pKa = 10.08RR718 pKa = 11.84DD719 pKa = 3.36IILNVDD725 pKa = 2.85TDD727 pKa = 4.04LFINRR732 pKa = 11.84NPITKK737 pKa = 9.74YY738 pKa = 9.09YY739 pKa = 8.36YY740 pKa = 10.45ARR742 pKa = 11.84EE743 pKa = 3.84IEE745 pKa = 4.04DD746 pKa = 3.4AKK748 pKa = 11.11RR749 pKa = 11.84EE750 pKa = 4.24GVVLPSKK757 pKa = 9.44PTGSQTRR764 pKa = 11.84ILGGDD769 pKa = 3.27KK770 pKa = 10.95VSS772 pKa = 3.38
Molecular weight: 88.8 kDa
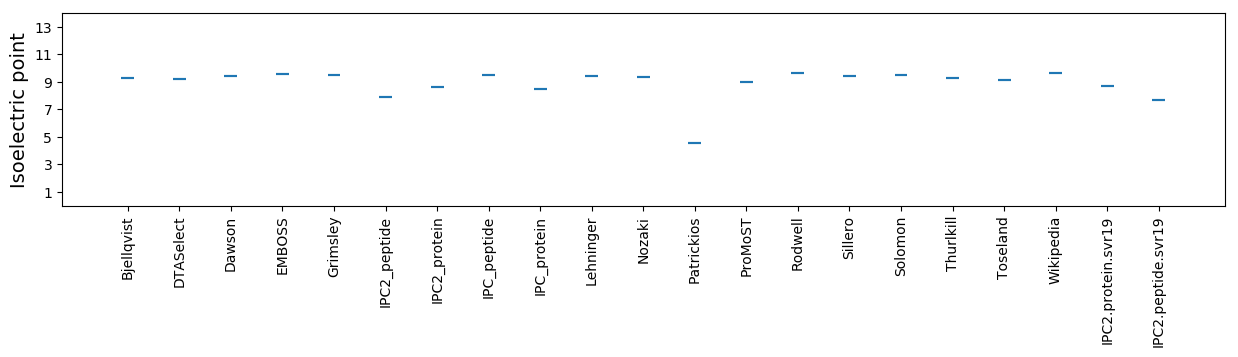
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191KCP0|A0A191KCP0_9VIRU RNA-dependent RNA polymerase OS=Cronartium ribicola mitovirus 5 OX=1816488 GN=RdRp PE=4 SV=1
MM1 pKa = 7.27SRR3 pKa = 11.84KK4 pKa = 8.92EE5 pKa = 3.97LYY7 pKa = 10.32LVNQSFKK14 pKa = 11.08SINNCSIIDD23 pKa = 3.83KK24 pKa = 10.58LKK26 pKa = 10.68LGPQVRR32 pKa = 11.84ILIWLAVKK40 pKa = 10.63VGLKK44 pKa = 10.11PSLSIQYY51 pKa = 7.94ITWFVTLTEE60 pKa = 4.06QVGIKK65 pKa = 9.42QAIQINKK72 pKa = 9.63SIRR75 pKa = 11.84LAFTRR80 pKa = 11.84WVCGDD85 pKa = 3.1TTIRR89 pKa = 11.84IEE91 pKa = 4.04KK92 pKa = 8.54VAVYY96 pKa = 10.61KK97 pKa = 10.78DD98 pKa = 4.19GYY100 pKa = 9.11PKK102 pKa = 10.56RR103 pKa = 11.84LRR105 pKa = 11.84QFRR108 pKa = 11.84NLIKK112 pKa = 9.92EE113 pKa = 4.17DD114 pKa = 4.16PSIKK118 pKa = 10.47SYY120 pKa = 11.09ILTLLNITRR129 pKa = 11.84SLEE132 pKa = 4.03LPLTLEE138 pKa = 4.42LSSVQDD144 pKa = 3.9PFTGLQTNIEE154 pKa = 4.4DD155 pKa = 3.97MYY157 pKa = 11.63KK158 pKa = 10.57DD159 pKa = 2.99IPKK162 pKa = 9.47FLNYY166 pKa = 9.23IRR168 pKa = 11.84SRR170 pKa = 11.84PGRR173 pKa = 11.84GKK175 pKa = 9.72IEE177 pKa = 4.07FKK179 pKa = 10.2KK180 pKa = 9.64WHH182 pKa = 6.01FTTKK186 pKa = 10.4NGPNGHH192 pKa = 7.4AYY194 pKa = 6.16TTMYY198 pKa = 10.79EE199 pKa = 4.09EE200 pKa = 5.56LRR202 pKa = 11.84TCSEE206 pKa = 4.54EE207 pKa = 3.94IIEE210 pKa = 4.71SISIVGGPKK219 pKa = 9.47IKK221 pKa = 10.61EE222 pKa = 4.07KK223 pKa = 10.84INDD226 pKa = 2.92IRR228 pKa = 11.84KK229 pKa = 9.25ILDD232 pKa = 3.8DD233 pKa = 3.65PTVYY237 pKa = 10.85SVMKK241 pKa = 10.33EE242 pKa = 3.92SLKK245 pKa = 10.71LKK247 pKa = 10.06EE248 pKa = 3.82ISEE251 pKa = 4.27PKK253 pKa = 9.05QAKK256 pKa = 8.33LTLIPDD262 pKa = 3.72KK263 pKa = 10.69EE264 pKa = 4.37GKK266 pKa = 8.08TRR268 pKa = 11.84VIGVGSYY275 pKa = 7.46WVQAGLKK282 pKa = 8.44PLHH285 pKa = 6.87DD286 pKa = 4.08FVMTVIRR293 pKa = 11.84RR294 pKa = 11.84IPGDD298 pKa = 3.19CTYY301 pKa = 10.47FQEE304 pKa = 4.53RR305 pKa = 11.84APKK308 pKa = 9.85ILRR311 pKa = 11.84KK312 pKa = 9.72QPGHH316 pKa = 6.59HH317 pKa = 6.06YY318 pKa = 10.2WSFDD322 pKa = 3.29LSSATDD328 pKa = 3.59RR329 pKa = 11.84FPRR332 pKa = 11.84VLQSKK337 pKa = 10.0IIASIYY343 pKa = 8.84NDD345 pKa = 3.89KK346 pKa = 10.78IATGWLTLISLPFHH360 pKa = 6.87FRR362 pKa = 11.84GQEE365 pKa = 3.41VKK367 pKa = 10.71YY368 pKa = 10.82AVGQPIGFYY377 pKa = 10.58SSWPVFTLTHH387 pKa = 6.85HH388 pKa = 5.6YY389 pKa = 9.52CVWVACKK396 pKa = 10.06RR397 pKa = 11.84AGVSPKK403 pKa = 10.5GVYY406 pKa = 10.25ALLGDD411 pKa = 5.48DD412 pKa = 5.13IVICHH417 pKa = 6.84DD418 pKa = 4.29LVAVKK423 pKa = 10.3YY424 pKa = 10.71LEE426 pKa = 4.91IISDD430 pKa = 4.13LGVDD434 pKa = 3.35ISKK437 pKa = 10.72QKK439 pKa = 6.43THH441 pKa = 6.95KK442 pKa = 10.79SSTCYY447 pKa = 10.19EE448 pKa = 4.18FAKK451 pKa = 10.12RR452 pKa = 11.84WYY454 pKa = 10.13QEE456 pKa = 3.97GDD458 pKa = 4.06SEE460 pKa = 4.33WSHH463 pKa = 6.26YY464 pKa = 9.27PLSGLRR470 pKa = 11.84PNAKK474 pKa = 8.79FTDD477 pKa = 3.96VLASTLQSRR486 pKa = 11.84SRR488 pKa = 11.84GWRR491 pKa = 11.84ISDD494 pKa = 3.77LTPVEE499 pKa = 3.71QASYY503 pKa = 10.46INCLIRR509 pKa = 11.84GWSSEE514 pKa = 3.78YY515 pKa = 10.37HH516 pKa = 5.98LSYY519 pKa = 10.29SINWFTVYY527 pKa = 9.07YY528 pKa = 9.95TSYY531 pKa = 10.62CAIRR535 pKa = 11.84DD536 pKa = 3.68FTTWMSPLRR545 pKa = 11.84EE546 pKa = 4.11LWRR549 pKa = 11.84ISPKK553 pKa = 10.02IQIVNPSYY561 pKa = 10.99DD562 pKa = 3.24HH563 pKa = 6.53PQAGEE568 pKa = 3.82HH569 pKa = 6.09FYY571 pKa = 10.92EE572 pKa = 4.81GLSARR577 pKa = 11.84VFALNGLSKK586 pKa = 10.58ILTASPKK593 pKa = 9.86ARR595 pKa = 11.84KK596 pKa = 9.45LFEE599 pKa = 4.58PLVKK603 pKa = 10.18FDD605 pKa = 4.1WYY607 pKa = 10.45FDD609 pKa = 4.61PIITDD614 pKa = 4.45DD615 pKa = 3.9EE616 pKa = 4.82NEE618 pKa = 4.16AQPLILPQVEE628 pKa = 4.07VLRR631 pKa = 11.84RR632 pKa = 11.84LNTSYY637 pKa = 11.08QSARR641 pKa = 11.84WEE643 pKa = 3.93EE644 pKa = 3.85LQGFFTGKK652 pKa = 10.36GPEE655 pKa = 4.11ANLSILVCDD664 pKa = 4.1ISTLFTTRR672 pKa = 11.84NQNLSWTVLNVLVKK686 pKa = 10.59EE687 pKa = 4.38MIKK690 pKa = 10.07VMKK693 pKa = 10.06IVRR696 pKa = 11.84HH697 pKa = 5.42TGFDD701 pKa = 3.66YY702 pKa = 11.09SSEE705 pKa = 4.28EE706 pKa = 4.02PLSSQLRR713 pKa = 11.84SAIYY717 pKa = 10.08RR718 pKa = 11.84DD719 pKa = 3.36IILNVDD725 pKa = 2.85TDD727 pKa = 4.04LFINRR732 pKa = 11.84NPITKK737 pKa = 9.74YY738 pKa = 9.09YY739 pKa = 8.36YY740 pKa = 10.45ARR742 pKa = 11.84EE743 pKa = 3.84IEE745 pKa = 4.04DD746 pKa = 3.4AKK748 pKa = 11.11RR749 pKa = 11.84EE750 pKa = 4.24GVVLPSKK757 pKa = 9.44PTGSQTRR764 pKa = 11.84ILGGDD769 pKa = 3.27KK770 pKa = 10.95VSS772 pKa = 3.38
MM1 pKa = 7.27SRR3 pKa = 11.84KK4 pKa = 8.92EE5 pKa = 3.97LYY7 pKa = 10.32LVNQSFKK14 pKa = 11.08SINNCSIIDD23 pKa = 3.83KK24 pKa = 10.58LKK26 pKa = 10.68LGPQVRR32 pKa = 11.84ILIWLAVKK40 pKa = 10.63VGLKK44 pKa = 10.11PSLSIQYY51 pKa = 7.94ITWFVTLTEE60 pKa = 4.06QVGIKK65 pKa = 9.42QAIQINKK72 pKa = 9.63SIRR75 pKa = 11.84LAFTRR80 pKa = 11.84WVCGDD85 pKa = 3.1TTIRR89 pKa = 11.84IEE91 pKa = 4.04KK92 pKa = 8.54VAVYY96 pKa = 10.61KK97 pKa = 10.78DD98 pKa = 4.19GYY100 pKa = 9.11PKK102 pKa = 10.56RR103 pKa = 11.84LRR105 pKa = 11.84QFRR108 pKa = 11.84NLIKK112 pKa = 9.92EE113 pKa = 4.17DD114 pKa = 4.16PSIKK118 pKa = 10.47SYY120 pKa = 11.09ILTLLNITRR129 pKa = 11.84SLEE132 pKa = 4.03LPLTLEE138 pKa = 4.42LSSVQDD144 pKa = 3.9PFTGLQTNIEE154 pKa = 4.4DD155 pKa = 3.97MYY157 pKa = 11.63KK158 pKa = 10.57DD159 pKa = 2.99IPKK162 pKa = 9.47FLNYY166 pKa = 9.23IRR168 pKa = 11.84SRR170 pKa = 11.84PGRR173 pKa = 11.84GKK175 pKa = 9.72IEE177 pKa = 4.07FKK179 pKa = 10.2KK180 pKa = 9.64WHH182 pKa = 6.01FTTKK186 pKa = 10.4NGPNGHH192 pKa = 7.4AYY194 pKa = 6.16TTMYY198 pKa = 10.79EE199 pKa = 4.09EE200 pKa = 5.56LRR202 pKa = 11.84TCSEE206 pKa = 4.54EE207 pKa = 3.94IIEE210 pKa = 4.71SISIVGGPKK219 pKa = 9.47IKK221 pKa = 10.61EE222 pKa = 4.07KK223 pKa = 10.84INDD226 pKa = 2.92IRR228 pKa = 11.84KK229 pKa = 9.25ILDD232 pKa = 3.8DD233 pKa = 3.65PTVYY237 pKa = 10.85SVMKK241 pKa = 10.33EE242 pKa = 3.92SLKK245 pKa = 10.71LKK247 pKa = 10.06EE248 pKa = 3.82ISEE251 pKa = 4.27PKK253 pKa = 9.05QAKK256 pKa = 8.33LTLIPDD262 pKa = 3.72KK263 pKa = 10.69EE264 pKa = 4.37GKK266 pKa = 8.08TRR268 pKa = 11.84VIGVGSYY275 pKa = 7.46WVQAGLKK282 pKa = 8.44PLHH285 pKa = 6.87DD286 pKa = 4.08FVMTVIRR293 pKa = 11.84RR294 pKa = 11.84IPGDD298 pKa = 3.19CTYY301 pKa = 10.47FQEE304 pKa = 4.53RR305 pKa = 11.84APKK308 pKa = 9.85ILRR311 pKa = 11.84KK312 pKa = 9.72QPGHH316 pKa = 6.59HH317 pKa = 6.06YY318 pKa = 10.2WSFDD322 pKa = 3.29LSSATDD328 pKa = 3.59RR329 pKa = 11.84FPRR332 pKa = 11.84VLQSKK337 pKa = 10.0IIASIYY343 pKa = 8.84NDD345 pKa = 3.89KK346 pKa = 10.78IATGWLTLISLPFHH360 pKa = 6.87FRR362 pKa = 11.84GQEE365 pKa = 3.41VKK367 pKa = 10.71YY368 pKa = 10.82AVGQPIGFYY377 pKa = 10.58SSWPVFTLTHH387 pKa = 6.85HH388 pKa = 5.6YY389 pKa = 9.52CVWVACKK396 pKa = 10.06RR397 pKa = 11.84AGVSPKK403 pKa = 10.5GVYY406 pKa = 10.25ALLGDD411 pKa = 5.48DD412 pKa = 5.13IVICHH417 pKa = 6.84DD418 pKa = 4.29LVAVKK423 pKa = 10.3YY424 pKa = 10.71LEE426 pKa = 4.91IISDD430 pKa = 4.13LGVDD434 pKa = 3.35ISKK437 pKa = 10.72QKK439 pKa = 6.43THH441 pKa = 6.95KK442 pKa = 10.79SSTCYY447 pKa = 10.19EE448 pKa = 4.18FAKK451 pKa = 10.12RR452 pKa = 11.84WYY454 pKa = 10.13QEE456 pKa = 3.97GDD458 pKa = 4.06SEE460 pKa = 4.33WSHH463 pKa = 6.26YY464 pKa = 9.27PLSGLRR470 pKa = 11.84PNAKK474 pKa = 8.79FTDD477 pKa = 3.96VLASTLQSRR486 pKa = 11.84SRR488 pKa = 11.84GWRR491 pKa = 11.84ISDD494 pKa = 3.77LTPVEE499 pKa = 3.71QASYY503 pKa = 10.46INCLIRR509 pKa = 11.84GWSSEE514 pKa = 3.78YY515 pKa = 10.37HH516 pKa = 5.98LSYY519 pKa = 10.29SINWFTVYY527 pKa = 9.07YY528 pKa = 9.95TSYY531 pKa = 10.62CAIRR535 pKa = 11.84DD536 pKa = 3.68FTTWMSPLRR545 pKa = 11.84EE546 pKa = 4.11LWRR549 pKa = 11.84ISPKK553 pKa = 10.02IQIVNPSYY561 pKa = 10.99DD562 pKa = 3.24HH563 pKa = 6.53PQAGEE568 pKa = 3.82HH569 pKa = 6.09FYY571 pKa = 10.92EE572 pKa = 4.81GLSARR577 pKa = 11.84VFALNGLSKK586 pKa = 10.58ILTASPKK593 pKa = 9.86ARR595 pKa = 11.84KK596 pKa = 9.45LFEE599 pKa = 4.58PLVKK603 pKa = 10.18FDD605 pKa = 4.1WYY607 pKa = 10.45FDD609 pKa = 4.61PIITDD614 pKa = 4.45DD615 pKa = 3.9EE616 pKa = 4.82NEE618 pKa = 4.16AQPLILPQVEE628 pKa = 4.07VLRR631 pKa = 11.84RR632 pKa = 11.84LNTSYY637 pKa = 11.08QSARR641 pKa = 11.84WEE643 pKa = 3.93EE644 pKa = 3.85LQGFFTGKK652 pKa = 10.36GPEE655 pKa = 4.11ANLSILVCDD664 pKa = 4.1ISTLFTTRR672 pKa = 11.84NQNLSWTVLNVLVKK686 pKa = 10.59EE687 pKa = 4.38MIKK690 pKa = 10.07VMKK693 pKa = 10.06IVRR696 pKa = 11.84HH697 pKa = 5.42TGFDD701 pKa = 3.66YY702 pKa = 11.09SSEE705 pKa = 4.28EE706 pKa = 4.02PLSSQLRR713 pKa = 11.84SAIYY717 pKa = 10.08RR718 pKa = 11.84DD719 pKa = 3.36IILNVDD725 pKa = 2.85TDD727 pKa = 4.04LFINRR732 pKa = 11.84NPITKK737 pKa = 9.74YY738 pKa = 9.09YY739 pKa = 8.36YY740 pKa = 10.45ARR742 pKa = 11.84EE743 pKa = 3.84IEE745 pKa = 4.04DD746 pKa = 3.4AKK748 pKa = 11.11RR749 pKa = 11.84EE750 pKa = 4.24GVVLPSKK757 pKa = 9.44PTGSQTRR764 pKa = 11.84ILGGDD769 pKa = 3.27KK770 pKa = 10.95VSS772 pKa = 3.38
Molecular weight: 88.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
772 |
772 |
772 |
772.0 |
88.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.145 ± 0.0 | 1.425 ± 0.0 |
4.793 ± 0.0 | 5.44 ± 0.0 |
3.886 ± 0.0 | 5.311 ± 0.0 |
1.943 ± 0.0 | 8.938 ± 0.0 |
7.383 ± 0.0 | 9.585 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.036 ± 0.0 | 3.368 ± 0.0 |
5.052 ± 0.0 | 3.756 ± 0.0 |
5.829 ± 0.0 | 8.42 ± 0.0 |
6.218 ± 0.0 | 6.218 ± 0.0 |
2.461 ± 0.0 | 4.793 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
