
Fulvivirga imtechensis AK7
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Fulvivirgaceae; Fulvivirga; Fulvivirga imtechensis
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
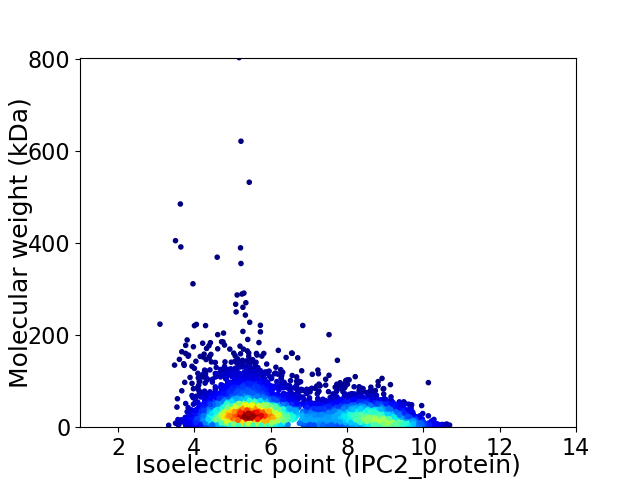
Virtual 2D-PAGE plot for 5952 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
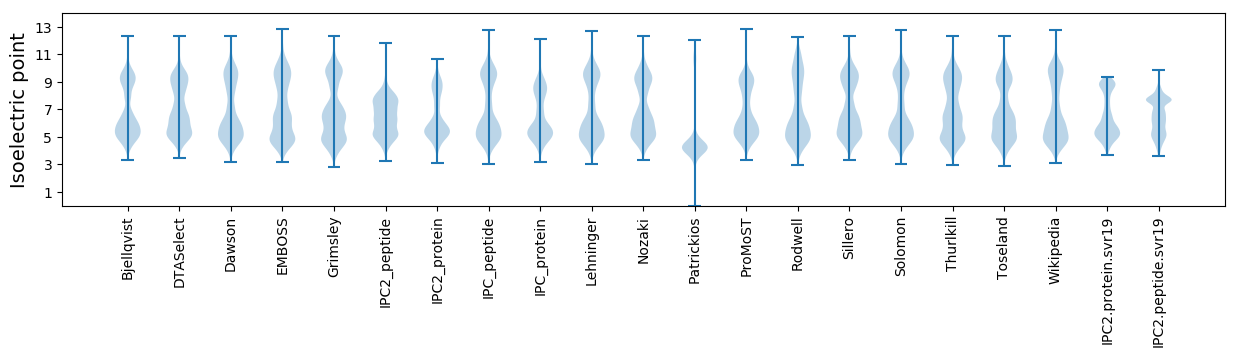
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8JI88|L8JI88_9BACT Uncharacterized protein OS=Fulvivirga imtechensis AK7 OX=1237149 GN=C900_00225 PE=4 SV=1
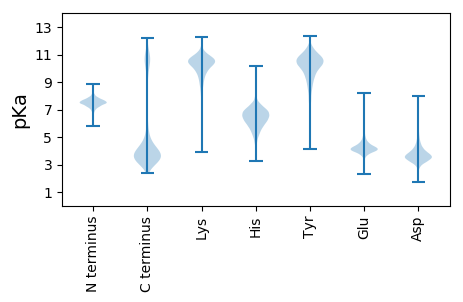
MM1 pKa = 7.51GNHH4 pKa = 5.84LQSLYY9 pKa = 10.81RR10 pKa = 11.84HH11 pKa = 6.04TLTKK15 pKa = 9.29YY16 pKa = 8.68KK17 pKa = 10.23KK18 pKa = 8.44YY19 pKa = 9.56RR20 pKa = 11.84SRR22 pKa = 11.84YY23 pKa = 7.97RR24 pKa = 11.84KK25 pKa = 9.96SIQTGLYY32 pKa = 8.85FQYY35 pKa = 10.41SQKK38 pKa = 10.17KK39 pKa = 7.71QRR41 pKa = 11.84LILEE45 pKa = 4.7RR46 pKa = 11.84IEE48 pKa = 4.26KK49 pKa = 10.13LKK51 pKa = 10.73QRR53 pKa = 11.84LASLKK58 pKa = 9.07WQIKK62 pKa = 7.43MAIASGALVMALHH75 pKa = 7.21AAPAQGQEE83 pKa = 3.93LGPFVEE89 pKa = 5.89APAKK93 pKa = 10.8NPFPPPPVSRR103 pKa = 11.84LTRR106 pKa = 11.84EE107 pKa = 3.64HH108 pKa = 6.77PAIVDD113 pKa = 3.21IDD115 pKa = 3.86NDD117 pKa = 3.41GDD119 pKa = 4.24LDD121 pKa = 4.03VFIGLTSGTIIFLKK135 pKa = 10.87NEE137 pKa = 3.69GSAEE141 pKa = 3.83KK142 pKa = 10.01PYY144 pKa = 10.51FVQQFSTDD152 pKa = 3.25NPANGIDD159 pKa = 3.37VGYY162 pKa = 10.41NATPTFADD170 pKa = 3.67LDD172 pKa = 4.01GDD174 pKa = 3.89GDD176 pKa = 4.0YY177 pKa = 11.62DD178 pKa = 5.61LIIGEE183 pKa = 4.24YY184 pKa = 9.82DD185 pKa = 3.16QYY187 pKa = 11.54PYY189 pKa = 10.61TPYY192 pKa = 11.61ANINYY197 pKa = 9.55YY198 pKa = 10.52EE199 pKa = 4.15NTGTASAPVFTLPSGEE215 pKa = 4.36HH216 pKa = 6.07PMDD219 pKa = 3.58NVFSDD224 pKa = 3.52SFYY227 pKa = 10.57GHH229 pKa = 7.4PAFVDD234 pKa = 3.03IDD236 pKa = 3.82NDD238 pKa = 3.4GDD240 pKa = 4.22LDD242 pKa = 4.13VFVGGARR249 pKa = 11.84NTIYY253 pKa = 9.83ATYY256 pKa = 10.66RR257 pKa = 11.84EE258 pKa = 4.53TIKK261 pKa = 10.47FWRR264 pKa = 11.84NDD266 pKa = 2.57GSLPVAQFTAEE277 pKa = 3.96EE278 pKa = 4.57GASNLISIVNPEE290 pKa = 3.94FGGTLDD296 pKa = 3.27HH297 pKa = 6.99AAPVLVDD304 pKa = 3.56IDD306 pKa = 4.07GDD308 pKa = 3.76GDD310 pKa = 3.44KK311 pKa = 11.62DD312 pKa = 3.77MFVGDD317 pKa = 3.7INGDD321 pKa = 2.76IRR323 pKa = 11.84FFRR326 pKa = 11.84NDD328 pKa = 2.79GTAEE332 pKa = 4.18SPSFPAEE339 pKa = 3.6ITGTGNPFDD348 pKa = 4.9GLTAPSFASPEE359 pKa = 4.03FGDD362 pKa = 4.22FDD364 pKa = 4.81NDD366 pKa = 3.21GDD368 pKa = 3.72FDD370 pKa = 6.23AIVGGGSSDD379 pKa = 3.66HH380 pKa = 5.98IQYY383 pKa = 10.49FEE385 pKa = 4.74NIGTASAPQFVEE397 pKa = 4.12RR398 pKa = 11.84FGPLNPFQGVDD409 pKa = 3.33VGYY412 pKa = 10.09HH413 pKa = 4.63STPAFVDD420 pKa = 2.98IDD422 pKa = 3.79NDD424 pKa = 3.59GDD426 pKa = 3.73LDD428 pKa = 3.89AFIGGKK434 pKa = 9.88YY435 pKa = 9.86SGYY438 pKa = 8.06NTGFVRR444 pKa = 11.84FFEE447 pKa = 4.37NTNGTFQEE455 pKa = 4.49KK456 pKa = 10.61NSTEE460 pKa = 4.03NPVEE464 pKa = 4.06GLSNVNFQNPSFVDD478 pKa = 3.03IDD480 pKa = 3.95GDD482 pKa = 3.94DD483 pKa = 4.04DD484 pKa = 3.75QDD486 pKa = 3.74YY487 pKa = 10.74FLDD490 pKa = 3.6IDD492 pKa = 4.17YY493 pKa = 10.89DD494 pKa = 3.52ILFYY498 pKa = 11.25EE499 pKa = 4.3NTGNASSMEE508 pKa = 4.18LGSANSSLLTPDD520 pKa = 4.3LEE522 pKa = 4.93GIVYY526 pKa = 10.26NYY528 pKa = 7.55KK529 pKa = 8.33TTFADD534 pKa = 3.11VDD536 pKa = 3.74RR537 pKa = 11.84DD538 pKa = 3.52GDD540 pKa = 3.72LDD542 pKa = 5.11AIGAHH547 pKa = 5.64TGYY550 pKa = 10.01PGGNLKK556 pKa = 10.52YY557 pKa = 10.11YY558 pKa = 10.8KK559 pKa = 9.5NTGTVNSPVFTLDD572 pKa = 3.57DD573 pKa = 4.46LNNPFSTLTEE583 pKa = 4.07TDD585 pKa = 3.4LSYY588 pKa = 11.4SPTPEE593 pKa = 4.38FVDD596 pKa = 3.34IDD598 pKa = 4.39HH599 pKa = 7.04NGHH602 pKa = 6.8ADD604 pKa = 3.62LFVGRR609 pKa = 11.84NTGTILFFPNTGAEE623 pKa = 4.14SFDD626 pKa = 3.78ISMTSANNPLAAVNVSSDD644 pKa = 3.45AAPAFADD651 pKa = 3.27IDD653 pKa = 4.08GDD655 pKa = 3.93GDD657 pKa = 3.82LDD659 pKa = 3.88AFIGSFSGRR668 pKa = 11.84ITYY671 pKa = 9.59FEE673 pKa = 4.41NQNQPPEE680 pKa = 3.85IVTNVGSALSFTEE693 pKa = 3.7NDD695 pKa = 3.28APLILDD701 pKa = 3.79AGFAISDD708 pKa = 4.64DD709 pKa = 3.95GDD711 pKa = 4.11DD712 pKa = 5.07DD713 pKa = 4.84IISAEE718 pKa = 4.09VAITANYY725 pKa = 9.59INGEE729 pKa = 4.22DD730 pKa = 3.73VLTFTPQGGVTGSFNASTGVLSLSGYY756 pKa = 10.75AKK758 pKa = 10.15LAVYY762 pKa = 9.79EE763 pKa = 4.72LVLQSITYY771 pKa = 9.74EE772 pKa = 4.09NTSEE776 pKa = 4.2NPSPAPRR783 pKa = 11.84TIEE786 pKa = 3.64LTAIDD791 pKa = 5.04FDD793 pKa = 4.1ATNPTSATSMTVNVIPVNDD812 pKa = 4.15APVLTATGSNVTYY825 pKa = 10.57TEE827 pKa = 4.58DD828 pKa = 4.46DD829 pKa = 3.89DD830 pKa = 5.18PGVVIDD836 pKa = 4.53PGVIISDD843 pKa = 3.57VDD845 pKa = 3.34NTTLSGATIAITANFVDD862 pKa = 4.39TEE864 pKa = 4.31DD865 pKa = 3.88VLDD868 pKa = 3.99FTDD871 pKa = 3.69QNGITGNYY879 pKa = 8.69DD880 pKa = 2.94AATGVLTLSGSATLAQYY897 pKa = 9.1EE898 pKa = 4.03AALRR902 pKa = 11.84SIAYY906 pKa = 9.87RR907 pKa = 11.84NTSQNPDD914 pKa = 3.32PATRR918 pKa = 11.84TVTFEE923 pKa = 4.31VNDD926 pKa = 3.88GTDD929 pKa = 3.35PSNTGTATVAVVPVNDD945 pKa = 4.05APVISSNNGTTATDD959 pKa = 4.17YY960 pKa = 10.96YY961 pKa = 11.11VEE963 pKa = 4.32EE964 pKa = 4.57SPVIIDD970 pKa = 3.61NLITISDD977 pKa = 3.71VDD979 pKa = 3.98DD980 pKa = 4.03TDD982 pKa = 4.93LEE984 pKa = 4.84AGEE987 pKa = 4.27VRR989 pKa = 11.84ITDD992 pKa = 4.16FVAGDD997 pKa = 3.58VLHH1000 pKa = 6.58FTDD1003 pKa = 3.82QNGITGSYY1011 pKa = 8.04DD1012 pKa = 3.3TQTGILALSGTATIEE1027 pKa = 4.0NYY1029 pKa = 7.34TTALQSITFEE1039 pKa = 4.41NNNAASGQRR1048 pKa = 11.84QIEE1051 pKa = 4.44FIINDD1056 pKa = 3.73GNADD1060 pKa = 3.52SSPYY1064 pKa = 8.7IRR1066 pKa = 11.84NMNVIANEE1074 pKa = 4.06PPVVDD1079 pKa = 3.93TTPQSTQVGNIVTIDD1094 pKa = 3.72LCDD1097 pKa = 5.38LISDD1101 pKa = 4.33PDD1103 pKa = 3.54NTYY1106 pKa = 10.92SEE1108 pKa = 4.27LTITVVSITSGAATDD1123 pKa = 3.65ISDD1126 pKa = 5.13CILTINYY1133 pKa = 8.94GSTGFSGQDD1142 pKa = 3.13QLVLQACDD1150 pKa = 3.43PSGACDD1156 pKa = 3.19QNTINIQVDD1165 pKa = 4.38EE1166 pKa = 4.47PSGPLTIYY1174 pKa = 10.77NAVSPNGDD1182 pKa = 3.61GLNDD1186 pKa = 2.57WWEE1189 pKa = 4.29IVNLTSPNKK1198 pKa = 8.95IALYY1202 pKa = 10.11NRR1204 pKa = 11.84WGDD1207 pKa = 3.73LVKK1210 pKa = 10.1TLTDD1214 pKa = 3.97YY1215 pKa = 11.42AGHH1218 pKa = 6.42TPNTLLNDD1226 pKa = 4.36LPAGTYY1232 pKa = 9.11YY1233 pKa = 11.12YY1234 pKa = 10.36KK1235 pKa = 10.42IKK1237 pKa = 10.63SPQGDD1242 pKa = 3.81YY1243 pKa = 11.14EE1244 pKa = 4.8GYY1246 pKa = 10.82LVIKK1250 pKa = 10.16KK1251 pKa = 10.14
MM1 pKa = 7.51GNHH4 pKa = 5.84LQSLYY9 pKa = 10.81RR10 pKa = 11.84HH11 pKa = 6.04TLTKK15 pKa = 9.29YY16 pKa = 8.68KK17 pKa = 10.23KK18 pKa = 8.44YY19 pKa = 9.56RR20 pKa = 11.84SRR22 pKa = 11.84YY23 pKa = 7.97RR24 pKa = 11.84KK25 pKa = 9.96SIQTGLYY32 pKa = 8.85FQYY35 pKa = 10.41SQKK38 pKa = 10.17KK39 pKa = 7.71QRR41 pKa = 11.84LILEE45 pKa = 4.7RR46 pKa = 11.84IEE48 pKa = 4.26KK49 pKa = 10.13LKK51 pKa = 10.73QRR53 pKa = 11.84LASLKK58 pKa = 9.07WQIKK62 pKa = 7.43MAIASGALVMALHH75 pKa = 7.21AAPAQGQEE83 pKa = 3.93LGPFVEE89 pKa = 5.89APAKK93 pKa = 10.8NPFPPPPVSRR103 pKa = 11.84LTRR106 pKa = 11.84EE107 pKa = 3.64HH108 pKa = 6.77PAIVDD113 pKa = 3.21IDD115 pKa = 3.86NDD117 pKa = 3.41GDD119 pKa = 4.24LDD121 pKa = 4.03VFIGLTSGTIIFLKK135 pKa = 10.87NEE137 pKa = 3.69GSAEE141 pKa = 3.83KK142 pKa = 10.01PYY144 pKa = 10.51FVQQFSTDD152 pKa = 3.25NPANGIDD159 pKa = 3.37VGYY162 pKa = 10.41NATPTFADD170 pKa = 3.67LDD172 pKa = 4.01GDD174 pKa = 3.89GDD176 pKa = 4.0YY177 pKa = 11.62DD178 pKa = 5.61LIIGEE183 pKa = 4.24YY184 pKa = 9.82DD185 pKa = 3.16QYY187 pKa = 11.54PYY189 pKa = 10.61TPYY192 pKa = 11.61ANINYY197 pKa = 9.55YY198 pKa = 10.52EE199 pKa = 4.15NTGTASAPVFTLPSGEE215 pKa = 4.36HH216 pKa = 6.07PMDD219 pKa = 3.58NVFSDD224 pKa = 3.52SFYY227 pKa = 10.57GHH229 pKa = 7.4PAFVDD234 pKa = 3.03IDD236 pKa = 3.82NDD238 pKa = 3.4GDD240 pKa = 4.22LDD242 pKa = 4.13VFVGGARR249 pKa = 11.84NTIYY253 pKa = 9.83ATYY256 pKa = 10.66RR257 pKa = 11.84EE258 pKa = 4.53TIKK261 pKa = 10.47FWRR264 pKa = 11.84NDD266 pKa = 2.57GSLPVAQFTAEE277 pKa = 3.96EE278 pKa = 4.57GASNLISIVNPEE290 pKa = 3.94FGGTLDD296 pKa = 3.27HH297 pKa = 6.99AAPVLVDD304 pKa = 3.56IDD306 pKa = 4.07GDD308 pKa = 3.76GDD310 pKa = 3.44KK311 pKa = 11.62DD312 pKa = 3.77MFVGDD317 pKa = 3.7INGDD321 pKa = 2.76IRR323 pKa = 11.84FFRR326 pKa = 11.84NDD328 pKa = 2.79GTAEE332 pKa = 4.18SPSFPAEE339 pKa = 3.6ITGTGNPFDD348 pKa = 4.9GLTAPSFASPEE359 pKa = 4.03FGDD362 pKa = 4.22FDD364 pKa = 4.81NDD366 pKa = 3.21GDD368 pKa = 3.72FDD370 pKa = 6.23AIVGGGSSDD379 pKa = 3.66HH380 pKa = 5.98IQYY383 pKa = 10.49FEE385 pKa = 4.74NIGTASAPQFVEE397 pKa = 4.12RR398 pKa = 11.84FGPLNPFQGVDD409 pKa = 3.33VGYY412 pKa = 10.09HH413 pKa = 4.63STPAFVDD420 pKa = 2.98IDD422 pKa = 3.79NDD424 pKa = 3.59GDD426 pKa = 3.73LDD428 pKa = 3.89AFIGGKK434 pKa = 9.88YY435 pKa = 9.86SGYY438 pKa = 8.06NTGFVRR444 pKa = 11.84FFEE447 pKa = 4.37NTNGTFQEE455 pKa = 4.49KK456 pKa = 10.61NSTEE460 pKa = 4.03NPVEE464 pKa = 4.06GLSNVNFQNPSFVDD478 pKa = 3.03IDD480 pKa = 3.95GDD482 pKa = 3.94DD483 pKa = 4.04DD484 pKa = 3.75QDD486 pKa = 3.74YY487 pKa = 10.74FLDD490 pKa = 3.6IDD492 pKa = 4.17YY493 pKa = 10.89DD494 pKa = 3.52ILFYY498 pKa = 11.25EE499 pKa = 4.3NTGNASSMEE508 pKa = 4.18LGSANSSLLTPDD520 pKa = 4.3LEE522 pKa = 4.93GIVYY526 pKa = 10.26NYY528 pKa = 7.55KK529 pKa = 8.33TTFADD534 pKa = 3.11VDD536 pKa = 3.74RR537 pKa = 11.84DD538 pKa = 3.52GDD540 pKa = 3.72LDD542 pKa = 5.11AIGAHH547 pKa = 5.64TGYY550 pKa = 10.01PGGNLKK556 pKa = 10.52YY557 pKa = 10.11YY558 pKa = 10.8KK559 pKa = 9.5NTGTVNSPVFTLDD572 pKa = 3.57DD573 pKa = 4.46LNNPFSTLTEE583 pKa = 4.07TDD585 pKa = 3.4LSYY588 pKa = 11.4SPTPEE593 pKa = 4.38FVDD596 pKa = 3.34IDD598 pKa = 4.39HH599 pKa = 7.04NGHH602 pKa = 6.8ADD604 pKa = 3.62LFVGRR609 pKa = 11.84NTGTILFFPNTGAEE623 pKa = 4.14SFDD626 pKa = 3.78ISMTSANNPLAAVNVSSDD644 pKa = 3.45AAPAFADD651 pKa = 3.27IDD653 pKa = 4.08GDD655 pKa = 3.93GDD657 pKa = 3.82LDD659 pKa = 3.88AFIGSFSGRR668 pKa = 11.84ITYY671 pKa = 9.59FEE673 pKa = 4.41NQNQPPEE680 pKa = 3.85IVTNVGSALSFTEE693 pKa = 3.7NDD695 pKa = 3.28APLILDD701 pKa = 3.79AGFAISDD708 pKa = 4.64DD709 pKa = 3.95GDD711 pKa = 4.11DD712 pKa = 5.07DD713 pKa = 4.84IISAEE718 pKa = 4.09VAITANYY725 pKa = 9.59INGEE729 pKa = 4.22DD730 pKa = 3.73VLTFTPQGGVTGSFNASTGVLSLSGYY756 pKa = 10.75AKK758 pKa = 10.15LAVYY762 pKa = 9.79EE763 pKa = 4.72LVLQSITYY771 pKa = 9.74EE772 pKa = 4.09NTSEE776 pKa = 4.2NPSPAPRR783 pKa = 11.84TIEE786 pKa = 3.64LTAIDD791 pKa = 5.04FDD793 pKa = 4.1ATNPTSATSMTVNVIPVNDD812 pKa = 4.15APVLTATGSNVTYY825 pKa = 10.57TEE827 pKa = 4.58DD828 pKa = 4.46DD829 pKa = 3.89DD830 pKa = 5.18PGVVIDD836 pKa = 4.53PGVIISDD843 pKa = 3.57VDD845 pKa = 3.34NTTLSGATIAITANFVDD862 pKa = 4.39TEE864 pKa = 4.31DD865 pKa = 3.88VLDD868 pKa = 3.99FTDD871 pKa = 3.69QNGITGNYY879 pKa = 8.69DD880 pKa = 2.94AATGVLTLSGSATLAQYY897 pKa = 9.1EE898 pKa = 4.03AALRR902 pKa = 11.84SIAYY906 pKa = 9.87RR907 pKa = 11.84NTSQNPDD914 pKa = 3.32PATRR918 pKa = 11.84TVTFEE923 pKa = 4.31VNDD926 pKa = 3.88GTDD929 pKa = 3.35PSNTGTATVAVVPVNDD945 pKa = 4.05APVISSNNGTTATDD959 pKa = 4.17YY960 pKa = 10.96YY961 pKa = 11.11VEE963 pKa = 4.32EE964 pKa = 4.57SPVIIDD970 pKa = 3.61NLITISDD977 pKa = 3.71VDD979 pKa = 3.98DD980 pKa = 4.03TDD982 pKa = 4.93LEE984 pKa = 4.84AGEE987 pKa = 4.27VRR989 pKa = 11.84ITDD992 pKa = 4.16FVAGDD997 pKa = 3.58VLHH1000 pKa = 6.58FTDD1003 pKa = 3.82QNGITGSYY1011 pKa = 8.04DD1012 pKa = 3.3TQTGILALSGTATIEE1027 pKa = 4.0NYY1029 pKa = 7.34TTALQSITFEE1039 pKa = 4.41NNNAASGQRR1048 pKa = 11.84QIEE1051 pKa = 4.44FIINDD1056 pKa = 3.73GNADD1060 pKa = 3.52SSPYY1064 pKa = 8.7IRR1066 pKa = 11.84NMNVIANEE1074 pKa = 4.06PPVVDD1079 pKa = 3.93TTPQSTQVGNIVTIDD1094 pKa = 3.72LCDD1097 pKa = 5.38LISDD1101 pKa = 4.33PDD1103 pKa = 3.54NTYY1106 pKa = 10.92SEE1108 pKa = 4.27LTITVVSITSGAATDD1123 pKa = 3.65ISDD1126 pKa = 5.13CILTINYY1133 pKa = 8.94GSTGFSGQDD1142 pKa = 3.13QLVLQACDD1150 pKa = 3.43PSGACDD1156 pKa = 3.19QNTINIQVDD1165 pKa = 4.38EE1166 pKa = 4.47PSGPLTIYY1174 pKa = 10.77NAVSPNGDD1182 pKa = 3.61GLNDD1186 pKa = 2.57WWEE1189 pKa = 4.29IVNLTSPNKK1198 pKa = 8.95IALYY1202 pKa = 10.11NRR1204 pKa = 11.84WGDD1207 pKa = 3.73LVKK1210 pKa = 10.1TLTDD1214 pKa = 3.97YY1215 pKa = 11.42AGHH1218 pKa = 6.42TPNTLLNDD1226 pKa = 4.36LPAGTYY1232 pKa = 9.11YY1233 pKa = 11.12YY1234 pKa = 10.36KK1235 pKa = 10.42IKK1237 pKa = 10.63SPQGDD1242 pKa = 3.81YY1243 pKa = 11.14EE1244 pKa = 4.8GYY1246 pKa = 10.82LVIKK1250 pKa = 10.16KK1251 pKa = 10.14
Molecular weight: 134.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8JSF3|L8JSF3_9BACT Uncharacterized protein OS=Fulvivirga imtechensis AK7 OX=1237149 GN=C900_02145 PE=4 SV=1
MM1 pKa = 7.08QVDD4 pKa = 3.5IFDD7 pKa = 5.93AILSDD12 pKa = 3.91SVYY15 pKa = 10.94LEE17 pKa = 4.06KK18 pKa = 10.55FLNQMHH24 pKa = 6.47SKK26 pKa = 8.91YY27 pKa = 10.39PPMHH31 pKa = 6.81AMRR34 pKa = 11.84ARR36 pKa = 11.84MMRR39 pKa = 11.84RR40 pKa = 11.84MYY42 pKa = 10.42RR43 pKa = 11.84FSEE46 pKa = 4.04VDD48 pKa = 3.34SLVIADD54 pKa = 3.63TLIRR58 pKa = 11.84RR59 pKa = 11.84NMLKK63 pKa = 10.79LMLNQADD70 pKa = 4.12RR71 pKa = 11.84DD72 pKa = 4.46SVFCRR77 pKa = 11.84QMSQSIMKK85 pKa = 9.65HH86 pKa = 4.68RR87 pKa = 11.84PLRR90 pKa = 11.84KK91 pKa = 9.16RR92 pKa = 11.84LRR94 pKa = 11.84QHH96 pKa = 5.85MGVPEE101 pKa = 3.87
MM1 pKa = 7.08QVDD4 pKa = 3.5IFDD7 pKa = 5.93AILSDD12 pKa = 3.91SVYY15 pKa = 10.94LEE17 pKa = 4.06KK18 pKa = 10.55FLNQMHH24 pKa = 6.47SKK26 pKa = 8.91YY27 pKa = 10.39PPMHH31 pKa = 6.81AMRR34 pKa = 11.84ARR36 pKa = 11.84MMRR39 pKa = 11.84RR40 pKa = 11.84MYY42 pKa = 10.42RR43 pKa = 11.84FSEE46 pKa = 4.04VDD48 pKa = 3.34SLVIADD54 pKa = 3.63TLIRR58 pKa = 11.84RR59 pKa = 11.84NMLKK63 pKa = 10.79LMLNQADD70 pKa = 4.12RR71 pKa = 11.84DD72 pKa = 4.46SVFCRR77 pKa = 11.84QMSQSIMKK85 pKa = 9.65HH86 pKa = 4.68RR87 pKa = 11.84PLRR90 pKa = 11.84KK91 pKa = 9.16RR92 pKa = 11.84LRR94 pKa = 11.84QHH96 pKa = 5.85MGVPEE101 pKa = 3.87
Molecular weight: 12.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1920940 |
37 |
7274 |
322.7 |
36.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.857 ± 0.029 | 0.762 ± 0.011 |
5.665 ± 0.023 | 6.783 ± 0.03 |
4.835 ± 0.029 | 6.855 ± 0.034 |
2.0 ± 0.016 | 7.336 ± 0.031 |
6.728 ± 0.045 | 9.409 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.282 ± 0.016 | 5.3 ± 0.029 |
3.625 ± 0.024 | 3.543 ± 0.02 |
4.211 ± 0.022 | 6.496 ± 0.031 |
5.45 ± 0.042 | 6.536 ± 0.027 |
1.134 ± 0.011 | 4.192 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
